Abstract
The complete mitochondrial genome of Dermestes ater DeGeer was obtained by high-throughput sequencing method. The genome was a circular molecule with a length of 15,810 bp and an average sequencing depth of 2375.9×, which contains 13 protein-coding genes (PCGS), 22 transfer RNA genes (tRNA), 2 ribosomal RNA genes (rRNA), and 1 control region. The overall base composition of the mitochondrial DNA was: A 40.85%, C 15.02%, G 10.04%, and T 34.09%. The phylogenetic analysis upon complete mitogenomics revealed that D. ater DeGeer is most closely related to Dermestes tessellatocollis Motschulsky, with high bootstrap values.
Introduction
Dermestes ater DeGeer (Coleoptera: Dermestidae) is a world-wide pest with important economic significance and mainly damages raw skin, dried fish, bacon, silk cocoon, raw silk, leather garment, woolen goods, and so on (Masayoshi Citation1970; Shibata et al. Citation1984). The present domestic and international studies of mitochondrial genome on Dermestidae is very limited, especially for D.ater, which mainly focused on biological characteristics (Roth and Willis Citation1950; Coombs Citation1981; Li et al. Citation2015; Duan, Wu, Liu, et al. Citation2018; Duan, Wu, Zheng, et al. Citation2018) and prevention (Chowdhary Citation1989; Ye Citation2007). In this study, the complete mitochondrial genome sequence of D. ater was determined by high-throughput sequencing method with Illumina Hiseq 2500 (Tsingke, Tianjin). Enriching the data of the mitochondrial genome, which laid a foundation for the molecular identification and genetic evolution of populations of the Dermatidae insect.
Specimen (voucher no. SZHT0523G142) was collected from the cocoon warehouse of Nanning, Guangxi Province, China (Latitude: 23.06349N, Longitude: 108.29758E) on August 2019, and stored in Sericulture Research Institute of College of Agricultural, Guangxi University (Nanning, China). The complete mitochondrial genome sequence was submitted to GenBank with accession number of MT742293.
In total, 10.94 G of raw data and 10.38 G of clean data were obtained for sequence by SPAdes version 3.9 (Tamura and Nei Citation1993). The whole mitochondrial genome of D. ater is 15,810 bp in length and constitutive of 38 genes. These genes contain 13 protein-coding genes (ATP6, ATP8, COI, COII, COIII, ND1, ND2, ND3, ND4, ND4L, ND5, ND6, and Cytb), 22 transfer RNA genes (tRNA), 2 ribosomal RNA genes (rRNA), and 2 control regions (D-loop). The base content was 40.85% A, 10.04% G, 34.09% T, and 15.02% C. The 65.87% of (A + T) showed great preference to AT.
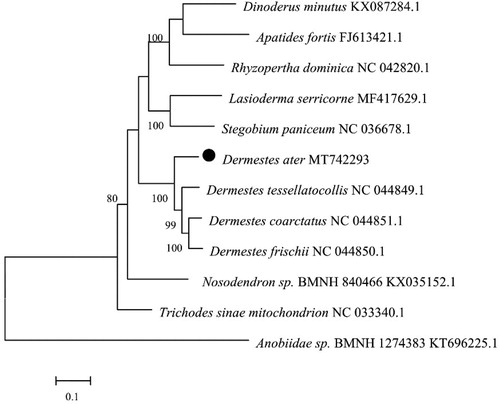
To investigate the phylogenetic relationship of D. ater, phylogenetic tree was constructed with other 11 species by MEGA7.0 that using the maximum-likelihood (ML) method to infer their taxonomic status (Kumar et al. Citation2016). The results showed that mtDNA of D. ater had a close relationship with that of the Dermestes tessellatocollis Motschulsky (). However, the molecular evidence inferred in this study were limited, further elucidations of the evolutionary relationships within D. ater, the more information on the mitochondrial genome of Coleoptera insects is needed.
Figure 1. Phylogenetic tree of the complete mitogenome of 12 species in Coleoptera. The complete mitochondrial genome was downloaded from GenBank and the phylogenic tree was constructed by neighbor-Joining method with 1000 bootstrap replicates. The gene’s accession number for tree construction is listed behind the species name.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank BankIt in NCBI at [https://www.ncbi.nlm.nih.gov/], reference number MT742293. Data are ethically correct to do and does not violate the protection of human subjects, or other valid ethical, privacy, or security concerns.
Additional information
Funding
References
- Chowdhary SK. 1989. Killing method of a large number of Dermestes ater Degeer in silkworm cocoon. Sericologia. 29(1):93–96.
- Coombs CW. 1981. The development, fecundity and longevity of Dermestes ater Degeer (Coleoptera: Dermestidae). Pergamon. 17(1):31–36.
- Duan YB, Wu HP, Liu JY, Dong ZS, Li J, Zheng XL, Lu W. 2018. Observation on antennal sensilla of Dermestes ater DeGeer with scaning electron microscope. Plant Protect. 44(01):95–100.
- Duan YB, Wu HP, Zheng XL, Li J, Lu W. 2018. Supercooling points and freezing points at different stage of Dermestes ater DeGeer. Plant Protect. 44(02):116–121.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li J, Zhuo QH, Huang LF, Liang X, Qu DC. 2015. The study of morphological characteristics and high temperature resistance of Dermestes ater Degeer. J Guangxi Agric. 30(02):22–24.
- Roth LM, Willis ER. 1950. The oviposition of dermestes ater DeGeer, with notes on bionomics under laboratory conditions. Am Midland Nat. 44(2):427.
- Masayoshi S. 1970. Attack of Dermestes ater (DEGEER) on the silkworm. Jpn Soc Sericult Sci. 39(3):201–203.
- Shibata M, Ito N, Ishitani A, Matsuda Y, Nishi K. 1984. Black larder beetles (Dermestes ater Degeer) and their pupal shells in a mummy with reference to the postmortem interval. Nihon Hoigaku Zasshi. 38(4):445–453.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10(3):512–526.
- Ye CY. 2007. Investigation report on the harm of moth to silkworm cocoon and its control method. China Fiber Inspect. (12):45–47.
