Abstract
Wiesnerella denudata (Mitt.) Steph. is a thallose liverwort distributed in Asian subtropical to tropical regions. It is the only one species in genus Wiesnerella and family Wiesnerellaceae. To investigate intraspecific variations on mitochondrial genomes of W. denudata, we completed mitochondrial genome of W. denudata. Its length is 185,640 bp, longer than that of the previously sequenced mitochondrial genome by 71 bp and contains 73 genes (41 protein-coding genes, 3 rRNAs, 28 tRNAs, and 1 pseudogene). A total of 149 single nucleotide polymorphisms (SNPs) and 3,033 insertions and deletions are identified, much higher than those of Marchantia polymorpha subsp. ruderalis and Riccia fluitans. Phylogenetic trees show that W. denudata is clustered with Monosolenium tenerum belonging to Monosoleniaceae.
Wiesnerlla denudata (Mitt.) Steph. was described from the East Himalayas by Mitten as Dumortiera denudata (Mitten Citation1860). Later Stephani transferred D. denudata to W. denudata (Stephani Citation1898). Inoue described a new family Wiesnerellaceae based on the monotypic genus with one species by four to six irregular valves of capsule dehiscence and specialized asexual structure absent (Inoue Citation1976). As such, this species has changed in the genus and the family. Recent phylogenetic study shows that Wienerellaceae was related to Ricciaceae (Crandall-Stotler et al. Citation2009). Whole organelle genome sequences will provide better resolution for uncovering phylogenetic relationship. We completed mitochondrial genome sequences of W. denudata for understanding its phylogenetic position.
The thallus of W. denudata collected in Jeju city, Korea (Voucher specimen in Jeonbuk National University Herbarium (JNU), Korea; S.S. Choi, CS-201126; 33.48318N, 126.73896E) was used for extracting DNA with DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using NovaSeq6000 at Macrogen Inc., Korea. Mitochondrial genome was completed by Velvet v1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser version v1.12 (Zhao et al. Citation2011), BWA version v0.7.17 (Li Citation2013), and SAMtools version v1.9 (Li et al. Citation2009) under the environment of Genome Information System (GeIS; http://geis.infoboss.co.kr; Park et al., in preparation). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on W. denudata mitochondrial genome (MK230933; Dong et al. Citation2019).
The mitochondrial genome of W. denudata (GenBank accession is MT745951) is 185,640 bp, which is longer than the previously sequenced mitochondrial genome (MK230933) by 71 bp. It contains 73 genes (41 protein-coding genes, 3 rRNAs, 28 tRNAs, and 1 pseudogene) and overall GC content is 42.9%. Nad7 gene was revealed as pseudogene in both W. denudata mitochondrial genomes which is similar to the other Marchantiales species (Dong et al. Citation2019; Kwon et al. Citation2019a, Citation2019b; Min et al. Citation2020).
The previous mitochondrial genome contains 203 polymorphic sites and 1,174 not determined bases occupying 0.74% of mitochondrial genome. Except these bases, 149 single nucleotide polymorphisms (SNPs; 0.080%) and 3,033 insertions and deletions (INDELs; 1.62%) were identified. Number and ratio of identified sequence variations are much higher than those of Marchantia polymorpha subsp. ruderalis (7 SNPs; 0.0038%; Kwon et al. Citation2019b), Riccia fluitans (18 SNPs; 0.0097% and 19 INDELs; 0.010%; Min et al. Citation2020), Dumortiera hirsuta (12 SNPs; 0.0067% and 24 INDELs; 0.013%; Dong et al. Citation2019; Kwon et al. Citation2019a), and Monosolenium tenerum (14 SNPs; 0.0075% and 7 INDELs; 0.0037%; Dong et al. Citation2019). They are even higher than those of vascular plants, including Liriodendron tulifipera (365 SNPs; 0.066%; 2,117 INDELs; 0.38%; Park et al. Citation2019) and Arabidopsis thaliana (64 SNPs; 0.017% and 1,089 INDELs; 0.30%; Park et al., in preparation). However, those of Scapania ampliata (823 SNPs; 0.057% and 2,242 INDELs; 1.56%; Choi et al., under revision) display similar level.
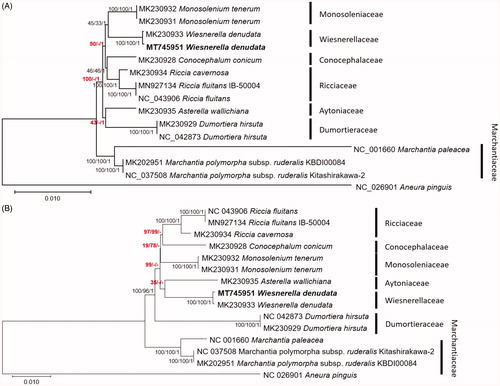
Fifteen complete mitochondrial genomes including two W. denudata mitochondrial genome were used for drawing neighbor-joining (bootstrap repeat is 10,000), maximum likelihood (bootstrap repeat is 1,000), and Bayesian Inference phylogenic trees (Number of generations is 1,100,000) with MEGA X (Kumar et al. Citation2018) and MrBayes v3.2.7a (Huelsenbeck and Ronquist Citation2001) based on alignments of both 37 conserved protein-coding genes based on annotation of the used mitochondrial genomes and whole mitochondrial genome sequences by MAFFT version v7.450 (Katoh and Standley Citation2013). Phylogenetic trees present that our W. denudata mitochondrial genome is clustered with the previous W. denudata mitogenome with high supportive values (), which is different from the previous study based on chloroplast genome (Choi et al. Citation2020). It also displays that Wiesnerellaceae is clustered with Monosoleniaceae, not Ricciaceae (), which is incongruent to the previous study (Crandall-Stotler et al. Citation2009). In addition, five nodes which cover higher level of taxa display incongruency of maximum likelihood tree or supportive values of maximum likelihood are lower than those of Bayesian Interference under the congruent between two trees (). In addition, phylogenetic trees constructed based on complete mitochondrial genome display more incongruent among three trees (). Maximum likelihood tree presents that Asterella wallichiana is clustered with W. denudata; while the rest two trees display that A. wallichiana is located as third basal group after Marchantiaceae and Dumortieraceae. These incongruencies of phylogenetic trees indicate that additional in-depth phylogenetic analyses should be conducted in near future.
Figure 1. Neighbor-joining (bootstrap repeat is 10,000), maximum likelihood (bootstrap repeat is 1000), and Bayesian Inference (Number of generations is 1,100,000) phylogenetic trees of (A) concatenation of alignment of conserved 37 genes (B) 15 complete mitochondrial genomes: Wiesnerella denudata (MT745951 in this study and MK230933; Dong et al. Citation2019), Monosolenium tenerum (MK230931 and MK230932; Dong et al. Citation2019), Conocephalum conicum (MK230928; Dong et al. Citation2019), Riccia cavernosa (MK230934; Dong et al. Citation2019), Riccia fluitans (NC_043906 and MN927134; Min et al. Citation2020), Asterella wallichiana (MK230935; Dong et al. Citation2019), Dumortiera hirsuta (NC_042873 and MK230929; Dong et al. Citation2019; Kwon et al. Citation2019a), Marchantia polymorpha subsp. ruderalis (NC_037508 and MK202951; Bowman et al. Citation2017; Kwon et al. Citation2019b), Marchantia paleacea (NC_001660; Oda et al. Citation1992), and Aneura pinguis (NC_026901; Myszczyński et al. Citation2017) as an outgroup. Gray bars with labels indicate specific classes. The numbers above branches indicate bootstrap support values of maximum likelihood, neighbor-joining, and Bayesian Inference phylogenetic trees, respectively. Phylogenetic trees were drawn based on maximum likelihood tree. Red-colored supportive values indicate incongruent among three trees.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number MT745951 in NCBI GenBank.
Additional information
Funding
References
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, et al. 2017. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 171(2):287–304.
- Choi SS, Kwon W, Park J. 2020. The complete chloroplast genome of Wiesnerella denudata (Mitt.) Steph. (Wiesnerellaceae, Marchantiophyta). Mitochondrial DNA Part B. 5(3):3142–3144.
- Crandall-Stotler B, Stotler R, Long D. 2009. Phylogeny and classification of the Marchantiophyta. Edinburgh J Bot. 66(1):155.
- Dong S, Zhao C, Zhang S, Zhang L, Wu H, Liu H, Zhu R, Jia Y, Goffinet B, Liu Y. 2019. Mitochondrial genomes of the early land plant lineage liverworts (Marchantiophyta): conserved genome structure, and ongoing low frequency recombination. BMC Genomics. 20(1):953.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Kwon W, Kim Y, Park J. 2019a. The complete mitochondrial genome of Dumortiera hirsuta (Sw.) Nees (Dumortieraceae, Marchantiophyta). Mitochondrial DNA Part B. 4(1):1586–1587.
- Kwon W, Kim Y, Park J. 2019b. The complete mitochondrial genome of Korean Marchantia polymorpha subsp. ruderalis Bischl. & Boisselier: inverted repeats on mitochondrial genome between Korean and Japanese isolates. Mitochondrial DNA Part B. 4(1):769–770.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997.
- Min J, Kwon W, Xi H, Park J. 2020. The complete mitochondrial genome of Riccia fluitans L.(Ricciaceae, Marchantiophyta): investigation of intraspecific variations on mitochondrial genomes of R. fluitans. Mitochondrial DNA Part B. 5(2):1220–1222.
- Inoue H. 1976. Illustrations of Japanese Hepaticae. Vol. 1. Tokyo: Tsukiji Shokan. p. 2–1. i–viii. p. 193.
- Mitten W. 1860. Hepaticae Indiae Orientalis: an enumeration of the Hepaticae of the East Indies. Bot J Linn Soc. 5(18):89–108.
- Myszczyński K, Bączkiewicz A, Szczecińska M, Buczkowska K, Kulik T, Sawicki J. 2017. The complete mitochondrial genome of the cryptic species C of Aneura pinguis. Mitochondrial DNA Part A. 28(1):112–113.
- Oda K, Yamato K, Ohta E, Nakamura Y, Takemura M, Nozato N, Akashi K, Kanegae T, Ogura Y, Kohchi T. 1992. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. A primitive form of plant mitochondrial genome. J Mol Biol. 223(1):1–7.
- Park J, Kim Y, Kwon M. 2019. The complete mitochondrial genome of tulip tree, Liriodendron tulipifera L. (Magnoliaceae): intra-species variations on mitochondrial genome. Mitochondrial DNA Part B. 4(1):1308–1309.
- Stephani F. 1898. Species Hepaticarum, continuation. Bull Herb Boissier. 7(5):381–407.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 12(14):S2.
