Abstract
Platycrater arguta is a rare and endangered shrub species endemic to East Asia. Here, we report the complete chloroplast (cp) genome structure and its taxonomic position within Hydrangeaceae to promote its conservation and restoration. The complete cp genome of P. arguta was 157,810 bp in length and contained a large single-copy region (LSC) of 86,823 bp and a small single-copy region (SSC) of 18,735 bp, as well as a pair of inverted repeat (IR) regions of 26,126 bp, each. 113 unique genes are predicted in this cp genome, including 79 protein-coding genes, 30 transfer RNA (tRNA) genes and 4 rRNAs. Maximum-likelihood (ML) phylogenetic analysis based on 79 shared cp CDS (coding DNA sequences) of 19 species reveals a close relationship between P. arguta and Schizophragma hydrangeoides.
Platycrater Sieb. et Zucc. is a monotypic genus in Hydrangeaceae that includes Platycrater arguta, a rare and endangered species endemic to East Asia (Fu Citation1989; Ao Citation2008). Studies on this rare and disjunct distributed temperate shrub are significant in understanding of the evolutionary history of Sino-Japanese Floristic Region (Qiu et al. Citation2009; Qi et al. Citation2014). Because of its small range size and small number of populations, the species has been considered ‘threatened’ by the China Species Red List (Wang and Xie Citation2004), and it was involved in the priority list of species of the greatest conservation concern in the Chinese Biodiversity Action Plan in 1994 (Ao Citation2008; Qiu et al. Citation2009). The classification of the Hydrangeaceae has long been problematic, and phylogenetic studies using a limited set of markers have often not been able to fully resolve relationships among this genus (Hufford et al. Citation2001). By taking advantages of next-generation sequencing technologies that efficiently provide the chloroplast (cp) genomic resources of our interested species, we can rapidly access the abundant genetic information for phylogenetic research and conservation genetics (Liu et al. Citation2017; Chen et al. Citation2019). Therefore, we sequenced the whole cp genome of P. arguta to elucidate its phylogenetic relationship within Hydrangeaceae.
Total genomic DNA was extracted from silica-dried leaves collected from Niutoushan, Linhai City (Zhejiang, China) using a modified CTAB method (Doyle and Doyle Citation1987). A voucher specimen (Li18001) was collected and deposited in the Herbarium of Taizhou University. Sequencing was conducted on the HiSeq 2500 platform. We assembled the cp genomes with NOVOPlasty (Dierckxsens et al. Citation2017) and annotated with the dual organellar genome annotator (DOGMA; Wyman et al. Citation2004). Then, BLAST was used to check the annotation, followed by manual correction through comparison with other closely related cp genomes of Hydrangeaceae in Geneious R11 (Biomatters Ltd., Auckland, New Zealand).
The complete cp genome of P. arguta (GenBank accession MT610904) was 157,810 bp long consisting of a pair of inverted repeat (IR) regions (26,126 bp, each) divided by large single-copy (LSC) and small single-copy regions (LSC) of 86,823 bp and 18,735 bp, respectively. The overall GC contents of the total length, LSC, SSC, and IR regions were 37.8, 36.0, 31.5, and 43.1%, respectively. A total of 113 unique genes were predicted and annotated, including 79 protein-coding genes, 30 transfer RNA (tRNA) genes, and four rRNAs. Among the 113 genes, three genes crossed adjacent regions: the ycf1 gene crossed the SSC/IRa junction, the rps19 gene crossed IRb/LSC junction, and ndhF crossed the IRb/SSC junction.
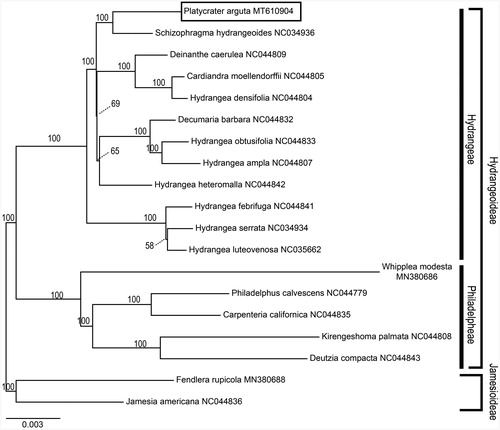
To investigate the taxonomic positions of P. arguta, a maximum-likelihood (ML) analysis was performed using RAxML-HPC2 version 7.6.3 (Stamatakis Citation2006) at the CIPRES Science Gateway (http://www.phylo.org/) using a cp CDS (coding DNA sequences) matrix consisting of CDS sequences of 19 Hydrangeaceae species and two outgroup taxa. The ML tree () was consistent with the most recent phylogenetic study on Hydrangeaceae (Soltis et al. Citation1995; Hufford et al. Citation2001). According to the phylogenetic tree, P. arguta belongs to tribe Hydrangeeae of subfamily Hydrangeoideae and closely related to Schizophragma hydrangeoides Sieb. et Zucc.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT610904. The raw sequencing reads used in this study were deposited in the Sequence Read Archive (SRA) under accession number PRJNA659226.
Additional information
Funding
References
- Ao CQ. 2008. Pre-zygotic embryological characters of Platycrater arguta, a rare and endangered species endemic to East Asia. J Plant Biol. 51(2):116–121.
- Chen LX, Brown KR, Yi XG, Sun ZS. 2019. Complete chloroplast genome of Prunus emarginata and its implications for the phylogenetic position within Prunus sensu lato (Rosaceae). Mitochondrial DNA Part B. 4(2):3402–3403.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Fu LG. 1989. Rare and endangered plants in China. Shanghai (China): Hanghai Education Publishing House.
- Hufford L, Moody ML, Soltis DE. 2001. A phylogenetic analysis of Hydrangeaceae based on sequences of the plastid gene matK and their combination with rbcL and morphological data. Int J Plant Sci. 162:835–846.
- Liu LX, Li R, Worth JRP, Li X, Li P, Cameron KM, Fu CX. 2017. The complete chloroplast genome of Chinese bayberry (Morella rubra, Myricaceae): implications for understanding the evolution of Fagales. Front Plant Sci. 8:968.
- Qi XS, Yuan N, Comes HP, Sakaguchi S, Qiu YX. 2014. A strong ‘filter’ effect of the East China Sea land bridge for East Asia’s temperate plant species: inferences from molecular phylogeography and ecological niche modelling of Platycrater arguta (Hydrangeaceae). BMC Evol Biol. 14(1):41.
- Qiu YX, Qi XS, Jin XF, Tao XY, Fu CX, Naiki A, Comes HP. 2009. Population genetic structure, phylogeography, and demographic history of Platycrater arguta (Hydrangeaceae) endemic to East China and South Japan, inferred from chloroplast DNA sequence variation. Taxon. 58:1226–1241.
- Soltis DE, Xiang QY, Hufford L. 1995. Relationships and evolution of Hydrangeaceae based on rbcL sequence data. Am J Bot. 82:504–514.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Wang S, Xie Y. 2004. China species red list. Vol. 1. Beijing (China): Higher Education Press.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.