Abstract
The total mitogenome of the confiscated material of tufted deer, Elaphodus cephalophus cephalophus was sequenced and annotated. It is 16,351bp in length and contained 2 ribosomal RNAs, 22 transfer RNAs (tRNAs), 13 protein-coding genes (PCGs) and, 1 control region (CR). Phylogenetic analyses showed the close relationship of Elaphodus and Muntiacus, and monophyletic clades of genera Elaphodus and Muntiacus.
Elaphodus cephalophus is a unique species of genus Elaphodus (monotypic) from a large area of southern China and northwestern Myanmar (Leslie et al. Citation2013) with three subspecies, E. c. cephalophus, E. c. ichangensis and E. c. michianus (Wang Citation2003). However, there are still controversies on the distribution of these subspecies. Among these three subspecies, E. c. cephalophus is distributed in western China (Wang Citation2003; Wang et al. Citation2007). Due to the overhunting and trading in remote areas, the species has been listed as ‘Near Threatened’’ by the IUCN (Harris and Jiang Citation2015). Despite available mitogenome of tufted deer (Pang et al. Citation2008; Zhang et al. Citation2019), reference data for the three subspecies mitogenome are still lacking at present. In this study, we determined and characterized the complete mitogenome of E. c. cephalophus and investigate the phylogenetics of the species with published homologous mitogenome genes of other related species.
The illegally traded tufted deer with only broken leg parts was confiscated material by local public security bureau for forest. The specimen studied here was collected from Dujiangyan (N30°59, E103°33), Sichuan Province, China, in 2019, which was deposited in the Museum of public security bureau of forest in Dujiangyan (Specimen voucher: DJY2019006). Species molecular identification was done by reference sequences of Elaphodus cephalophus retrieved from GenBank (Wang and Lan Citation2000; Pang et al. Citation2008). Subspecies affiliation (E. c. cephalophus) was identified according to geographic locality (poaching site) provided by local public security bureau of forest, in combination with known subspecies distribution of the three tufted deer subspecies (Wang Citation2003; Pan et al. Citation2007; Wang et al. Citation2007). The leg muscle tissue was preserved in absolute ethanol and stored in −80 °C refrigerator. DNA from muscle tissue was isolated using the Ezup pillar genomic DNA extraction kit (Sangon Biotech, Shanghai, China). The mitogenome was amplified in 11 overlapping segments by PCR with 11 sets of primers designed by Pang et al (Pang et al. Citation2008).
The total mitogenome sequence of E. c. cephalophus (16,351 bp, GenBank accession number: MT726046) contains 13 protein coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, 22 tRNA genes, and a control region (D-loop). The total nucleotide composition of the E. c. cephalophus mitogenome is A (33.3%), G (13.4%), C (23.9%) and T (29.4%). Most genes are encoded on heavy strand (H strand), except the nad6 and eight tRNAs (tRNA-P, Q, A, N, C, Y, S2, and E) are located on the L-light strand (L strand). Nine of the protein coding genes (PCGs) began with ATG, except nad2, nad3, and nad5 with ATT, and nad4l with GTG. Three PCGs (nad2, atp8 and cytb) ended with TAG, and six (nad1, cox1, cox2, atp6, nad4l, nad5 and nad6) terminated by TAA. For the incomplete stop codon, two PCGs (cox3 and nad3) ended with TA, whereas nad4 stopped with a single base T. 12S and 16S rRNA are 957 and 1566 bp in length, respectively.
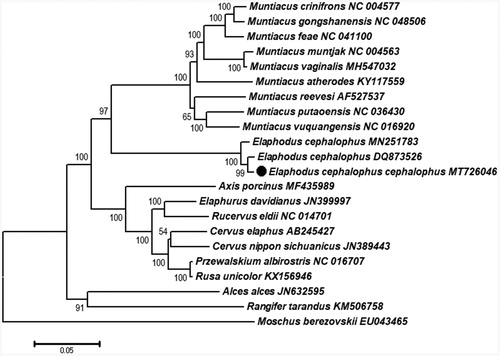
Phylogenetic analysis was performed using 11 genera (Rusa, Przewalskium, Cervus, Rucervus, Elaphurus, Axis, Elaphodus, Muntiacus, Rangifer, Alces, and Moschus) based on combined PCGs. The Maximum likelihood tree was reconstructed by MEGA7.0 (Kumar et al. Citation2016) with Moschus berezovskii as outgroup (). The newly sequenced E. c. cephalophus (Dujiangyan population) and E. cephalophus (DQ873526) from unknown locality formed a well-supported clade that was the sister clade to the Yingjing county population (MN251783). Our results also indicated that the geographic differentiation corresponds to the mitochondrial preliminary differences of these populations. Genus Elaphodus was closely related to the Muntiacus, and the genera (Elaphodus and Muntiacus) are monophyletic with high bootstrap support. The mitogenome data of subspecies E. c. cephalophus will benefit the three subspecies identification and support future effective conservation and management strategies plans by authorities.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of this study are available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MT726046, reference number MT726046.
References
- Harris R, Jiang Z. 2015. Elaphodus cephalophus. The IUCN Red List of Threatened Species. 2015eT7112A22159620.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Leslie D, Lee D, Dolman R. 2013. Elaphodus cephalophus (Artiodactyla: Cervidae). BioOne. 45(904):80–91.
- Pan Q, Wang Y, Kun Y. 2007. A field guide to the mammals of China. Beijing: China Forestry Publishing House.
- Pang H, Liu W, Chen Y, Fang L, Zhang X, Cao X. 2008. Identification of complete mitochondrial genome of the tufted deer. Mitochondrial DNA. 19(4):411–417.
- Wang Y. 2003. A complete checklist of mammal species and subspecies in china: a taxonomic and geographic reference. Chengdu: China Forestry Publishing House.
- Wang W, Lan H. 2000. Rapid and parallel chromosomal number reductions in muntjac deer inferred from mitochondrial DNA phylogeny. Mol Biol Evol. 17(9):1326–1333.
- Wang W, Xia L, Hu J. 2007. Differentiation of cranial characteristics of three subspecies of tufted deer in China. Sichuan J Zool. 26:777–781.
- Zhang L, Yao Y, Li D, Wu J, Wen A, Xie M, Wang Q, Zhu G, Ni Q, Zhang M, et al. 2019. Complete mitochondrial genome sequence of tufted deer (Elaphodus cephalophus) with phylogenetic analysis. Mitochondr DNA Part B. 4(2):3778–3779.