Abstract
This article describes the complete chloroplast genome of Gnetum luofuense. The G. luofense plastome was 114,795 bp in length, containing a large single copy region (66,103 bp) and a small single copy region (9438 bp), separated by two inverted repeat regions (19,627 bp). The genome lost all ndh genes and contained 116 genes, including 68 protein-coding genes, 40 tRNA genes, and eight rRNA genes. The GC content was 33.3%, 12 genes all contained an intron, ycf3 gene contained two introns while rps12 was a transpliced gene. Phylogenetic analysis using 61 concatenated protein-coding genes suggests that G. luofuense with the rest of other gnetophytes were sister to or nested within all conifers.
Gnetales is a small group of gymnosperms comprising three genera: Gnetum, Ephedra, and Welwitschia. Among extant seed plants, the phylogenetic placement of Gnetales remains controversial (Ran et al. Citation2018). Thus, various hypotheses regarding its exact taxonomic position were proposed. This includes the ‘gnepine’ (sister to pines), ‘gnetifer’ (sister to all conifers), ‘gnecup’ (sister to non-Pinaceae cupressophytes), and ‘anthophyte’ (sister to angiosperms) hypotheses (Zhong et al. Citation2010). Gnetum luofuense C.Y. Cheng is one of the lianoid species of Gnetum distributed in the South Eastern (SE) part of China. Morphologically, it is differentiated from other species of the genus by the presence of 9–11 involucral collars in male spikes and coarsely reticulate-wrinkled seeds (dried) (Fu et al. Citation1999). Currently, it is classified as Near Threatened (NT) by the IUCN Red List of Threatened Species (Baloch Citation2011; IUCN Citation2020). Chloroplast genomes, herewith referred to as plastomes, are often used in phylogenetic research because they are highly conserved and less complex compared to nuclear and mitochondrial DNA but still hold sufficient genomic data despite having relatively smaller dimensions (Chang et al. Citation2020). At present, six completely sequenced chloroplast genomes under the genus Gnetum were deposited in NCBI GenBank and the addition of G. luofuense aimed to contribute additional sequence data for future studies resolving issues in Gnetales phylogeny and seed plants evolution.
Fresh leaves of G. luofuense were collected from Sanzhaigu, Yonghan, Longmen County, Guangdong, China (Chen et al. 2018123101, SZG, N29°35′30.9′′, E113°56′2.1′′). DNA was extracted using SDS method (Shi et al. Citation2012) and stored at −80 °C prior to sequencing using Illumina HiSeq 2500 platform. Paired-end sequences with an average read length of 150 bp were generated with an average sequencing depth of 2164.6X. After filtering reads, the final circular plastome was de novo assembled using SPAdes version 3.11.0 (Vasilinetc et al. Citation2015) and was annotated using CpGAVAS (Liu et al. Citation2012), RNAmmer version 1.2 Server (Lagesen et al. Citation2007), and tRNAscan-SE (Chan and Lowe Citation2019) for protein-coding genes, rRNA and tRNA annotations, respectively. The complete nucleotide sequence of G. luofuense plastome was deposited in NCBI GenBank with accession number MT663149.
The plastome of G. luofuense has a circular quadripartite structure with a total genome length of 114,795 bp. It contained a large single copy region (66,103 bp) and a small single copy region (9438 bp), separated by two inverted repeat regions (19,627 bp). The genome lost all ndh genes and contained 116 genes, including 68 protein-coding genes, 40 tRNA genes, and eight rRNA genes. The GC content of G. luofuense plastome is 33.3%. The genes trnK-UUU, trnG-UCC, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, rpl2, trnI-GAU, and trnA-UGC all contained one intron, the ycf3 gene contained two introns while rps12 gene was found to be transpliced.
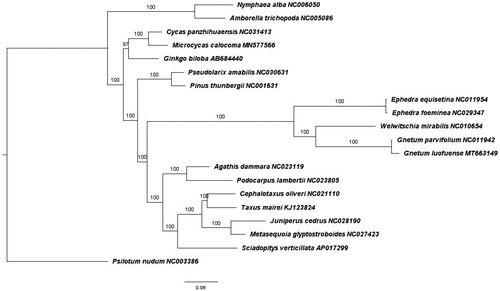
To determine the phylogenetic position of G. luofuense within seed plants, the complete plastome sequence was aligned with other 19 species representing basal angiosperms and gymnosperms (Cycadales, Coniferales, and Ginkgoales). For Gnetales, at least two species per genus were selected for phylogenetic analysis with the exception of the monotypic Welwitschia. Maximum-likelihood tree () was constructed using GTR + Gamma substitution model and 1000 bootstrap replicates using built-in RAxML in CIPRES (Miller et al. Citation2010). Using Psilotum nudum (Psilotales) as outgroup, phylogenetic tree indicated that G. luofuense and Gnetum parvifolium were closest to the genus Welwitschia diverging and forming a distinct clade with Ephedra. Moreover, all three Gnetales genera were phylogenetically placed within all conifers, similar to the study conducted by Wan et al. (Citation2018). Hence, our chloroplast genome topology was closest to the ‘gnetifer’ hypothesis but not congruent with ‘gnepine,’ ‘gnecup,’ or ‘anthophyte’ hypotheses.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All chloroplast genome sequences generated and used in this study are accessible via NCBI GenBank database. The complete nucleotide sequence of G. luofuense plastome was deposited in NCBI GenBank with accession number MT663149 (https://www.ncbi.nlm.nih.gov/nuccore/MT663149).
Additional information
Funding
References
- Baloch E. 2011. Gnetum luofuense. The IUCN red list of threatened species. 2011:e.T194922A8919354. https://www.iucnredlist.org/species/194922/8919354. Downloaded on 22 June 2020.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Meth Mol Biol. 1962:1–14.
- Chang ACG, Lai Q, Chen T, Tu T, Wang Y, Agoo EMG, Duan J, Li N. 2020. The complete chloroplast genome of Microcycas calocoma (Miq.) A. DC. (Zamiaceae, Cycadales) and evolution in Cycadales. PeerJ. 8:e8305.
- Fu LG, Yu YF, Gilbert MG. 1999. Gnetaceae. Flora of China. Vol. 4. p. 102–105. Published on the Internet http://www.efloras.org [accessed 22 June 2020] Missouri Botanical Garden, St. Louis, MO & Harvard University Herbaria, Cambridge, MA.
- IUCN. 2020. The IUCN red list of threatened species. Version 2020-2. https://www.iucnredlist.org. Downloaded on 22 June 2020.
- Lagesen K, Hallin PF, Rodland E, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35 (9):3100–3108.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13:715.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Piscataway (NJ): IEEE.
- Ran JH, Shen TT, Wang MM, Wang XQ. 2018. Phylogenomics resolves the deep phylogeny of seed plants and indicates partial convergent or homoplastic evolution between Gnetales and angiosperms. Proc Royal Soc B. 285:20181012.
- Shi C, Hu N, Huang H, Gao J, Zhao YJ, Gao LZ. 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS One. 7(2):e31468.
- Vasilinetc I, Prjibelski AD, Gurevich A, Korobeynikov A, Pevzner PA. 2015. Assembling short reads from jumping libraries with large insert sizes. Bioinformatics. 31 (20):3262–3268.
- Wan T, Liu ZM, Li LF, Leitch AR, Leitch IJ, Lohaus R, Liu ZJ, Xin HP, Gong YB, Liu Y, et al. 2018. A genome for gnetophytes and early evolution of seed plants. Nat Plants. 4(2):82–89.
- Zhong B, Yonezawa T, Zhong Y, Hasegawa M. 2010. The position of Gnetales among seed plants: overcoming pitfalls of chloroplast phylogenomics. Mol Biol Evol. 27(12):2855–2863.