Abstract
The complete chloroplast (cp) genome sequence of LHeucanthemella linearis was newly analyzed in this study. It was 151,395 bp in length and was a typical circular structure composed of a large single-copy region (LSC) (83,080 bp) and a small single-copy (SSC) region (18,391 bp) which were separated by two inverted repeat regions (24,962 bp). It was totally 6 bp shorter than the Chinese L. linearis cp genome and was composed of 132 genes. There were four regional specific Indels between them in the LSC region as well as 20 bp insertion in the intergenic spacer region excluding poly-A/T sequence variation. And it was clear that both of Luecanthemella linearis were sister to Chrysanthemum – Artemisia groups and their phylogenetic relationship from this study.
Luecanthemella linearis (Matsum. Ex Matsum.) Tzvelev is a member of the family Asteraceae and was treated as Chrysanthemum lineare Matsum. or Leucanthemum lineare (Matsum.) Vorosch. depending on the taxonomic recognition in the traditional classification. It is distributed in East Asia Manshuria, China, Korea, and Japan (El-Twab and Kondo Citation2007) against to the European native species L. serotina. Although just these two species comprise the genus Leucanthemella, they were scattered in the phylogenetic tree of Chrysanthemum and its related taxa (Masuda et al. Citation2009) and it needs comprehensive study for resolving this relationship. Luecanthemella linearis has generally large head flower with white ligulates and grows up to over 1 m. It is restricted inland marsh area with a less individual and was nominated as one of endangered species in the red data books of Korea due to its peculiar habitat and small population size (Korea National Arboretum Citation2009).
We obtained the plant material of Leucanthemella linearis which was collected from Mt. Chilbo-san of Suwon-si. Gyeonggi-do, Korea (37° 15.250′ N, 126° 56.060′ E, alt.204 M) and the voucher (CBNU2018-0242) was deposited at the Herbarium of Chungbuk National University (CBNU). Using the dried leaves samples, complete chloroplast (cp) genome of Leucanthemella linearis (MN883842) was sequenced by HiSeq4000 of Illumina and compared the sequence difference with Chinese L. linearis genome (NC043835). Totally 108,070,890 paired-end reads (2 × 151bp) were obtained and 100,955,336 reads were used for the assembly to the reference sequence after trimming with the length range 50–151 bp. The assembled reads were de novo assembled using the Geneious assembler. Using the assembled contigs, we conducted to align and repeat the procedure up to make a single contig. Complete cp genome was annotated using Geneious version 10.2.6 (Kearse et al. Citation2012) with manual correction and tRNAScan-SE (Lowe and Eddy Citation1997) for tRNA gene. The average coverage of this cp genome was 217.1×. The phylogenetic tree was constructed with related Chrysanthemum and Artemisia members and related Asteraceae based on the concatenated 78 coding genes using RAxML (Stamatakis Citation2014) after model test by ModelFinder (Kalyaanamoorthy et al. Citation2017).
The complete cp genome of Leucanthemella linearis has a typical circular structure with 151,395 bp in length and comprised a large single-copy region (LSC, 83,080 bp), a small single-copy region (SSC, 18,391 bp), and two inverted repeat regions (IR, 24,962 bp). It was totally 6 bp shorter than the Chinese L. linearis genome. The GC contents were 37.3%. It was composed of 132 genes and they were identified 87 coding genes, eight rRNA genes, and 37 tRNA genes. From the comparison of both L. linearis cp genomes, we found four regional specific Indels between them in the LSC region excluding poly-A/T sequence variation. They were 42 and 15 bp insertion on the present Korean L. linearis cp genome in trnK intron and intergenic spacer region of trnT-psbD, respectively, as well as 15 and 9 bp deletion in the intergenic spacer region of trnK-rps16 and petN-psbM. In the inverted repeat region, there was 20 bp insertion in the intergenic spacer region between ndhB and trnC and 1 bp deletion of poly-A in the SSC region of Korean L. linearis genome. These indels will be useful for developing the molecular markers to detect regional variation of Korean and Chinese species.
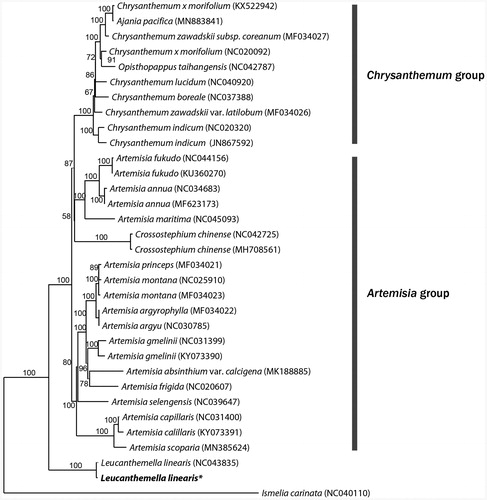
From the result of molecular phylogenetic analysis, both of Luecanthemella linearis were sister to Chrysanthemum – Artemisia groups and their phylogenetic relationship were highly supported (). It will provide more information for understanding the diversity and relationship of the family Asteraceae.
Acknowledgments
The authors thank to Dr. K Choi of the Korea National Arboretum and her colleague for their cooperation to collect the plant material for this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI database at https://www.ncbi.nlm.nih.gov/, reference number [MN883842] after this article is published.
Additional information
Funding
References
- El-Twab MHA, Kondo K. 2007. FISH physical mapping of 5S rDNA and telomere sequence repeats identified a peculiar chromosome mapping and mutation in Leucanthemella linearis and Nipponanthemum nipponicum in Chrysanthemum sensu lato. Chromosome Bot. 2(1):11–17.
- Kearse M, Moir R, Wilson A, Stone-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Korea National Arboretum. 2009. Rare plants data book of Korea. Pocheon-si, Gyeonggi-do, Korea: Korea National Arboretum; p. 132.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Masuda Y, Yukawa T, Kondo K. 2009. Molecular phylogenetic analysis of members of Chrysanthemum and its related genera in the tribe Anthemideae, the Asteraceae in East Asia on the basis of the internal transcribed spacer (ITS) region and the external transcribed spacer (ETS) region of nrDNA. Chromosome Bot. 4(2):25–36.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. Model finder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.