Abstract
Austinograea species are restricted to hydrothermal vents and are typically considered to be omnivorous predators in vent communities. Here we present the complete mitochondrial genome of Austinograea sp. which was collected from Carlsberg Ridge, the mid-ocean ridge in the northwestern Indian Ocean. The genome is 15,584 bp in length with a 68.11% AT content. It contains 13 protein-coding genes (PCGs), 2 ribosomal RNA genes, and 22 transfer RNA genes. Phylogenetic analysis shows that the present species is closest to Austinograea rodriguezensis. This study contributes to further phylogenetic analysis within Eubrachyura.
All identified species in the genus Austinograea except Austinograea rodriguezensis colonize hydrothermal vents in the western Pacific Ocean at present and the factors that drove the adaptation and speciation of Austinograea remain unknown (Hessler and Martin Citation1989; Guinot Citation1990; Tsuchida and Hashimoto Citation2002; Mateos et al. Citation2012; Kim et al. Citation2013; Guinot and Segonzac Citation2018). The examination of phylogenetic relationships among Austinograea species could help to clarify the interaction between biological evolution and geographical processes at deep-sea hydrothermal vents (Lee et al. Citation2019). In this study, the complete mitochondrial genome from a deep-sea Austinograea sp. was characterized, and its relationship with closely related species was investigated.
The specimen was collected from Carlsberg Ridge, the mid-ocean ridge in the northwestern Indian Ocean (60°31′48″E, 6°21′36″N, 2920 m depth) using the Chinese manned submersible Jiaolong, and was identified as Austinograea sp. based on its morphological characters. The specimen (SRSIO17030301) and its DNA (DNASIO17030301) are deposited in the Sample Repository of the Second Institute of Oceanography, Ministry of Natural Resources, Hangzhou, China. DNA was extracted with QIAamp Tissue Kit (QIAGEN, Hilden, Germany) and mitochondrial DNA was amplified with a DNA REPLI-g Mitochondrial DNA Kit (QIAGEN, Hilden, Germany) as directed by the manufacturer. Library construction and sequencing were performed by Biozeron (Biozeron, Shanghai, China) using the Illumina Hiseq4000 sequencing platform (Illumina, San Diego, CA).
The complete mitogenome sequence of Austinograea sp. is 15,584 bp in length with a 68.11% AT content. It contains 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. Among the 37 genes, both rRNA genes (rrnL and rrnS) are encoded on the light strand, as in the other crustacean mitochondrial genomes. Eight tRNA genes (trnC-tgc, trnF-ttc, trnH-cac, trnL1-cta, trnP-cca, trnQ-caa, trnV-gta, and trnY-tac) are encoded on the light strand. Only four protein-coding genes (PCGs) (nad1, nad4, nad4L, and nad5) are encoded on the light strand, whereas the other nine PCGs are located on the heavy strand. Eight PCGs (cox1, cox2, atp8, atp6, cox3, nad4, cob, and nad2) are initiated by ATG. Two PCGs (nad4L and nad6) are started by ATT. The other three PCGs (nad1, nad3, and nad5) are initiated by ATA, ATC, TTG, respectively. Ten PCGs (nad3, cox1, cox2, atp6, cox3, nad4, cob, nad2, nad4L, and nad6) terminate with the typical TAA as stop codon, while the remaining three PCGs (nad1, atp8, and nad5) end with TAG. A total of 22 transfer RNA genes range in size from 62 to 73 bp. The mitogenome of Austinograea sp. has been deposited in GenBank under accession number MT855983.
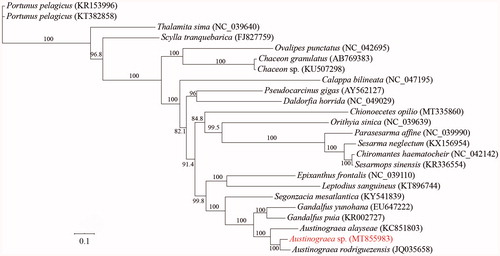
To reveal the phylogenetic relationship of Austinograea sp. with the other crabs, a total of 22 species with 23 mitochondrial genomes from Eubrachyura were obtained from the GenBank database. The mitochondrial PCGs were used for phylogenetic analysis by maximum likelihood (ML) method. In the phylogenetic tree, Austinograea sp. was closely related to A. rodriguezensis, and they clustered with A. alayseae into a branch. Meanwhile, Austinograea had a close relationship with Gandalfus in Bythograeoidea (). In this study, we present the complete mitochondrial genome sequence of Austinograea sp., which would contribute to further phylogenetic and comparative mitogenome studies of Eubrachyura. Furthermore, more mitochondrial genomic data of undetermined taxa and further analysis are required to reveal phylogeny and evolution of crabs.
Acknowledgments
The authors thank the Jiaolong team for their help in the specimen collection during the expedition.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All sequences generated or used in the present study are deposited in NCBI GenBank (www.ncbi.nlm.nih.gov/) and the accession numbers are detailed in .
Additional information
Funding
References
- Guinot D, Segonzac M. 2018. A review of the brachyuran deep-sea vent community of the western Pacific, with two new species of Austinograea Hessler & Martin, 1989 (Crustacea, Decapoda, Brachyura, Bythograeidae) from the Lau and North Fiji Back-Arc Basins. Zoosystema. 40(1):1–107.
- Guinot D. 1990. Austinograea alayseae sp. nov., crabe hydrothermal découvert dans le bassin de Lau, Pacifique sud-occidental (Crustacea Decapoda Brachyura). Bull Museum Natl D'hist Nat A (Zool). 11:879–903.
- Hessler RR, Martin JW. 1989. Austinograea williamsi, new genus, new species, a hydrothermal vent crab (Decapoda: Bythograeidae) from the Mariana Back-Arc Basin, western Pacific. J Crustacean Biol. 9(4):645–661.
- Kim SJ, Lee KY, Ju SJ. 2013. Nuclear mitochondrial pseudogenes in Austinograea alayseae hydrothermal vent crabs (Crustacea: Bythograeidae): effects on DNA barcoding. Mol Ecol Resour. 13(5):781–787.
- Lee WK, Kim SJ, Hou BK, Van Dover CL, Ju SJ. 2019. Population genetic differentiation of the hydrothermal vent crab Austinograea alayseae (Crustacea: Bythograeidae) in the Southwest Pacific Ocean. PLOS One. 14(4):e0215829.
- Mateos M, Hurtado LA, Santamaria CA, Leignel V, Guinot D. 2012. Molecular systematics of the deep-sea hydrothermal vent endemic brachyuran family Bythograeidae: a comparison of three Bayesian species tree methods. PLOS One. 7(3):e32066.
- Tsuchida S, Hashimoto J. 2002. A new species of bythograeid crab, Austinograea rodriguezensis (Decapoda, Brachyura), associated with active hydrothermal vents from the Indian Ocean. J Crustacean Biol. 22(3):642–650.