Abstract
Actinidia callosa var. strigillosa is one of the endangered Actinidia species scattering across mountain areas in southwest China. Here, we assembled and characterized the complete chloroplast (cp) genome sequence of A. strigillosa to provide genomic resources for its identification, conservation and utilization. The cp genome is 155,957 bp in length, containing a pair of 23,119 bp inverted repeat (IR) regions, which is separated by a large single copy region (LSC) of 89,429 bp and a small single copy region (SSC) of 20,290 bp. A total of 132 genes were annotated in this cp genome, including 83 protein-coding genes, 41 tRNA genes and 8 rRNA genes. The phylogenetic position of A. strigillosa based the cp genome data was sister to the group A. deliciosa and A. chinensis.
The genus Actinidia contains 54 species and 21 infraspecific taxa (Li et al. Citation2007). Currently, few species, such as A. chinensis Planchon and A. arguta (Siebold & Zuccarini) Planchon ex Miquel, were domesticated and commercialized (Ferguson Citation2004). Most taxa occurred naturally in China and many species have not been studied both in their genomic data and agronomic characteristics. Actinidia callosa var. strigillosa is one of the endangered species which distributed in Yunan, Guangxi, Guizhou, and Hunan province in China (Huang Citation2014). In this study, we reported complete sequence of chloroplast genome of A. strigillosa to provide basic genetic information to facilitate its future conservation and utilization.
The genomic DNA was extracted from leaves of A. strigillosa cultivated in National Kiwifruit Germplasm Genebank of China, Wuhan Botanical Garden, the Chinese Academy of Sciences (30°33′48.6″ N, 114°25′1.2″ E). An Illumina pair-end (PE) genomic library of 500-bp insert was constructed by following the PE standard protocol (Illumina, USA) and sequenced using an Illumina Hiseq 2000 platform by NovoGene (Beijing, China). The raw sequencing data were filtered and trimmed by fastp program (Chen et al. Citation2018), and then fed into NOVOPlasty (Dierckxsens et al. Citation2017) for assembly using cp genome of A. chinensis as the reference (Yao et al. Citation2015). The assembled genome sequence was then annotated using Plastid Genome Annotator (PGA) (Qu et al. Citation2019) against the cp genome of A. strigillosa. The annotation result was inspected using Geneious 11.0.4 software (Biomatters Limited, NZ; Matthew et al. Citation2012) and adjusted manually as needed. The sequence of chloroplast genomes was deposited in GenBank (accession numbers MT700484).
The circular cp genome of A. strigillosa was 155,957 bp (GC Content: 37.25%) in length, including a LSC region of 89,429 bp (GC Content: 35.51%), a SSC region of 20,290 bp (GC Content: 31.19%), and a pair of IR regions of 23,720 bp (GC Content: 43.27%). The GC content of A. strigillosa cp genomes is similar to the reported A. chinensis cp genomes (Yao et al. Citation2015). The cp genome of A. strigillosa encodes a total of 132 predicted functional genes, including 83 protein-coding genes, 41 tRNA genes, and 8 rRNA genes. Five protein-coding (i.e. rps7, ndhB, ycf15, ycf2, and rps12), eight tRNA (i.e. trnM-CAU, trnL-CAA, trnR-ACG, trnN-GUU, trnV-GAC, trnE-UUG, and trnA-UGC) and all four rRNA genes (rrn16, rrn23, rrn5, rrn4.5) are duplicated in the IR regions.
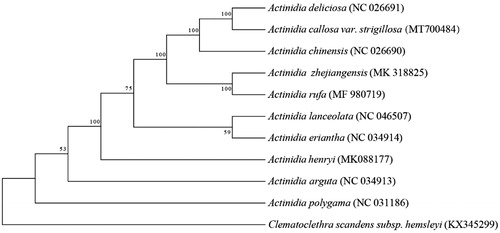
To identify the phylogenetic position of A. strigillosa, sequence alignment of eleven chloroplast genomes was conducted using MAFFT v.7 (Katoh and Standley Citation2013). The Maximum-Likelihood (ML) tree was constructed using MEGA 6.0 with 1000 bootstrap replicates (Swofford Citation2003). The chloroplast genome of A. strigillosa was shown to be closely related to that of A. deliciosa ().
Figure 1. Phylogenetic position of A. strigillosa in Actinidia as inferred by MP analyses of chloroplast genome sequences. Numbers above the lines indicate the maximum likelihood bootstrap value >50% for each clade. Accession Numbers: A. chinensis (NC026690), A. deliciosa (NC026691), A. zhejiangensis (MK318825), A. rufa (MF980719), A. lanceolata (NC046507), A. eriantha (NC034914), A. henryi (MK088177), A. arguta (NC034913), A. polygama (NC031186), and Clematoclethra scandens subsp. hemsleyi (KX345299).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank (accession numbers) at [https://www.ncbi.nlm.nih.gov/nuccore], reference number [MT700484].
Additional information
Funding
References
- Chen S, Yanqing Z, Yaru C, Jia G. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Ferguson AR. 2004. 1904–The year that kiwifruit (Actinidia deliciosa) came to New Zealand. New Zeal J Crop Hort. 32(1):3–27.
- Huang H. 2014. The genus Actinidia, a world monograph. Beijing, China: Science Press.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li JQ, Li XW, Soejarto DD. 2007. A revision of the genus Actinidia from China. Acta Hortic. 753(753):41–44.
- Matthew K, Richard M, Amy W, Steven SH, Matthew C, Shane S, Simon B, Alex C, Sidney M, Chris D. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (* and other methods), Version 4b 10. Sunderland (MA): Sinauer Associates.
- Yao X, Tang P, Li Z, Li D, Liu Y, Huang H. 2015. The first complete chloroplast genome sequences in Actinidiaceae: genome structure and comparative analysis. PLoS One. 10(6):e0129347.
