Abstract
Coelogyne fimbriata is an important orchid species with high medicinal value. Its complete chloroplast genome is 158,935 bp in length, which possesses the typical structure consising of a small single-copy region (SSC) of 18,743 bp, two inverted repeats (IRs) of 26,374 bp, and a large-single copy region (LSC) of 87,444 bp. The genome encodes 137 genes, including 91 protein-coding genes (PCGs), 38 tRNA genes and 8 rRNA genes. And the overall GC content is 37.40%. In addition, our phylogenetic analysis based on cp genome revealed the phylogenetic relationship between C. fimbriata and other 22 species in Orchidaceae.
Coelogyne fimbriata (Orchidaceae) is a perennial herb native to the forest edge at an altitude of 500-1200 m in southern Jiangxi, Guangdong, Hainan, Guangxi, Yunnan and Southeast Tibet, China (Chen et al. Citation1999). Coelogyne fimbriata has been reported to have important medicinal value (Chen et al. Citation2006; Sharma et al. Citation2017). However, the population resources of C. fimbriata continuously face decreasing due to the habitat destruction and transitional collection. Recently, Chloroplast DNA-based studies provide a remarkable breakthrough with data invaluable for studying genetic history and phylogeny (Yang et al. Citation2012; Givnish et al. Citation2015). Thus, we sequenced and characterized the plastome of C. fimbriata using the next-generation sequencing method to reveal the phylogenetic relationship of C. fimbriata.
We sampled the fresh leaf material from a plant specimen (now it is stored in the herbarium of Plant Biology Department, Beijing Forestry University, voucher No. Yachang 201901), which was collected from Leye County, Baise City, Guangxi Province, with geographic coordinates of 24.810152°N, 106.373218°E. We chose CTAB method to extract total genomic DNA (Doyle and Doyle Citation1987), and Illumina Novaseq 6000 platform was applied to perform 300 bp pair-end sequencing with the strategy of Illumina PE150. Plastome genome assembly was performed by Geneious (version 11, https://www.geneious.com), with the assembling algorithm of medium-low sensitivity. Due to the lack of published chloroplast genome sequences of the genus Coelogyne in NCBI database, clean reads were mapped to published chloroplast genome of some species (Orchidaceae): Pleione bulbocodioides (NC_036342), P. chunii (MK792342), P. formosana (NC_042197), P. forrestii (MK370035), Changnienia amoena (NC_045402), Dendrobium nobile (KX377961) and Dendrobium parciflorum (LC193512) as references. Filtered reads were then used for de novo assembly method with Geneious R11. The complete chloroplast sequence was annotated using PGA (Qu et al. Citation2019; Wang et al. Citation2020). The MAFFT software was implemented in multiple sequence alignment of full chloroplast sequences (Katoh and Standley Citation2013). Finally, we obtained a complete chloroplast genome of Coelogyne fimbriata and submitted it to GenBank. The accession number is MT548043.
The complete double-stranded circular chloroplast genome of Coelogyne fimbriata is 1,58,935 bp in length, containing a typical quadripartite structure consisting of two inverted repeats (IRs) of 26,374 bp each, separated by a large single-copy region (LSC) of 87,444 bp and a small single-copy (SSC) region of 18,743 bp. The total GC content is 37.40%. The plastome genome contains 137 functional genes, including 91 protein-coding genes (PCGs), 38 tRNA genes and 8 rRNA genes. Among them, 12 protein-coding genes (rps16, atpF, ropC1, ycf3, rps12, clpP, petB, petD, rpl16, rpl2, ndhB, rps12) and 6 tRNA genes (trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnI-GAU, trnA-UGC) have introns.
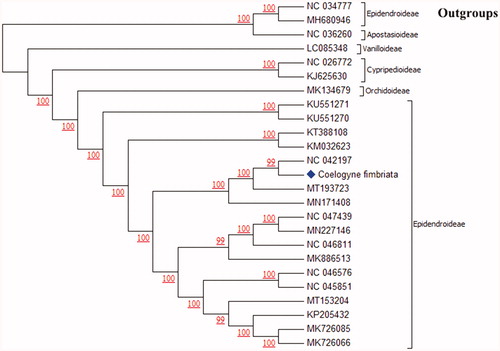
We constructed the phylogenetic tree based on 22 related complete chloroplast genomes of orchids and 2 non-orchid species downloaded from GenBank. All sequences were subsequently checked and unified gene alignment in Genious R11. The maximum-likelihood-based phylogenetic tree was constructed by MEGA (version 6, https://www.megasoftware.net/) under the nucleotide substitution model of Tamura-Nei, which has been evaluated as a more practical and advantageous model in estimating the branch length of the tree, especially when the number of nucleotides is very large (Tamura and Nei Citation1993). The results showed that Coelogyne fimbriata grouped with Pleione formosana (NC042197) with 99% bootstrap support (1000 replicates) (). Our phylogenetic tree shows agreements with the previous studies including morphologic and molecular identification. Many molecular-based phylogeny studies demonstrated that Coelogyne, Pleione, Bletilla and Arundina were grouped together (Burns-Balogh and Funk Citation1986; Givnish et al. Citation2015; Freudenstein and Chase Citation2015; Tang et al. Citation2017; Tan et al. Citation2020). Similar results were obtained by methods of the traditional morphological classification, such as Coelogyne and Pleione were belonged to the tribe of Coelogyninae (Chen et al. Citation1999, 2009). The cp sequence we published provides a reference for the discussion of the genetic diversity in Orchidaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The basic data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov/) with the accession number of MT548043, or available from the corresponding author.
Additional information
Funding
References
- Burns-Balogh P, Funk V. 1986. A phylogenetic analysis of the Orchidaceae. Smithsonian Contr Bot. 61(61):1–79.
- Chen SC, Liu ZJ, Zhu GH, Lang KY, Ji ZH, Luo YB, Jin XH, Cribb PJ, Wood JJ, Gale SW, et al. 2009. Orchidaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 25. St. Louis, Beijing: Science Press, Missouri Botanical Garden Press.
- Chen SC, Tsi ZH, Lang KY, Zhu GH. 1999. Trib. Epidendreae. In: Editorial Committee of Chinese Flora, Chinese Academy of Sciences, editors. Flora Republicae Popularis Sinicae. Vol. 18. Beijing: Science Press.
- Chen YG, Xu TP, Wu HY. 2006. Advance on the chemical and pharmacological studies on plants of the genus Coelogyne. Lishizhen Med Meter Med Res. 17:338–340.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Freudenstein JV, Chase MW. 2015. Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Ann Bot. 115(4):665–681.
- Givnish TJ, Spalink D, Ames M, Lyon SP, Hunter SJ, Zuluaga A, Iles WJD, Clements MA, Arroyo MTK, Leebens-Mack J, et al. 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc R Soc B. 282(1814):20151553.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. Pga: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Sharma C, Irshad S, Khatoon S, Arya KR. 2017. Pharmacognostical evaluation of Indian folk-traditional plants Coelogyne cristata and Pholidota articulata used for healing fractures. Indian J Exp Biol. 55(9):622–627.
- Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 10(3):512–526.
- Tan QQ, Li L, Zhang J, Wang YQ, Luo Y, Li QQ. 2020. Pollinia development in Arundina graminifolia (Orchidaceae) with taxonomic implications. Guihaia. 40(1):83–94.
- Tang H, Xiang L, Li XW, Sun W, Wang MN, Huang YF, Ye M. 2017. DNA barcoding identification of endangered medicinal plants of Orchidaceae. Chin J Chin Mater Med. 42(11):2058–2067.
- Wang TM, Kao HX, Cheng J. 2020. The complete chloroplast genome sequences of a wild diploid alfalfa Medicago edgeworthii (Leguminosae). Mitochondrial DNA B. 5(2):1683–1684.
- Yang HQ, Dong YR, Gu ZJ, Liang N, Yang JB. 2012. A preliminary assessment of matK, rbcL and trnH-psbA as DNA barcodes for Calamus (Arecaceae) species in China with a note on ITS. Ann Bot Fennici. 49(5–6):319–330.