Abstract
We sequenced the complete chloroplast genome of Zanthoxylum acanthopodium via high-throughput sequencing technology, and analyzed its structural characteristics and phylogenetic relationships. The chloroplast exhibits a genome length of 158,473 bp, including a pair of inverted repeat regions (IRa and IRb) of 27,369 bp, a small single-copy (SSC) region of 17,629 bp and a large single-copy (LSC) region of 85,570 bp. The annotation analysis identified 112 genes, containing 78 protein-coding genes, 30 tRNA genes and 4 rRNA genes. The phylogenetic analysis showed that Z. acanthopodium was closely related to Z. piperitum and Z. tragodes.
The Zanthoxylum genus belongs to the Rutaceae family. Plants in this genus are mainly utilized as spices and medicinal agents. About 250 species distributed in tropical and temperate regions of Asia and North America. In southern Yunnan and southeast Tibet of China, Zanthoxylum acanthopodium is a small tree with a height of 4.0 m, and grows in mountain shrubs or sparse forests at an altitude of 1,400–2,500 m (Editorial Committee of Chinese Journal of Plant of Chinese Academy of Sciences Citation1997). Its fruit is widely used as a spice or additional flavor for traditional delicacy. Z. acanthopodium was extensively used in traditional folk medicine and possess anti-inflammatory, antibacterial properties (Yanti et al. Citation2011; Majumder et al. Citation2014; Bhatt et al. Citation2018). However, there have been no genomic studies on Z. acanthopodium. Herein, we reported and characterized complete chloroplast genome of Z. acanthopodium, which will provide more sequences information to investigate the genome and phylogenetic relationship of the Rutaceae family.
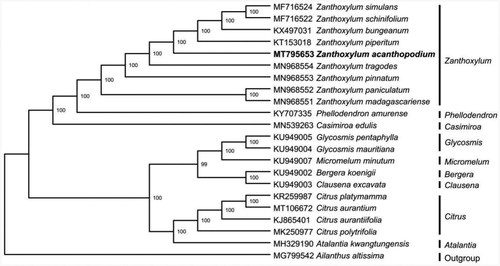
The experimental materials of Z. acanthopodium were collected from Dali county, Yunnan province, China (100°15′42.76″E, 25°31′33.38″N). The voucher herbarium specimen (No. ZSY111) were deposited at the Herbarium of Medicinal Plants and Crude Drugs of the College of Pharmacy and Chemistry, Dali University. High-quality genomic DNA was extracted using modified CTAB method (Doyle Citation1987; Yang et al. Citation2014). Whole-genome sequencing was conducted with pair-end (2 × 300 bp) library on Illumina Hiseq 2500 (Novogene, Tianjing, China) platform. To decrease the redundant data, the original reads were filtered by Trimmomatic v.0.32 software with default parameters (Bolger et al. Citation2014), the obtained clean reads were assembled into circular contigs using GetOrganelle.py (Jin et al. Citation2018) with Z. simulans (No. NC_037482) as the reference. Finally, the complete chloroplast genome sequences of Z. acanthopodium were annotated using Geneious R11.0.2 (Kearse et al. Citation2012), and then submitted to GenBank with the accession number of MT795653. To investigate its phylogenetic position within family Rutaceae, the complete chloroplast genome sequences of Z. acanthopodium and 20 related species from NCBI with Ailanthus altissima as an outgroup were aligned using MAFFT V.7.149 (Katoh and Standley Citation2013). Then, the maximum likelihood tree was generated by RAxML (Stamatakis Citation2014) with 1000 bootstrap replicates under the GTRGAMMAI substitution model.
The chloroplast genome of Z. acanthopodium was 1,58,473 bp in length with a typical quadripartite structure, containing a large single copy-region (LSC, 85,570 bp) and a small single copy-region (SSC, 17,625 bp), which were separated by two inverted repeat (IRa/IRb, 27,639 bp). The genome contained 112 genes, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The overall GC content of the genome is 38.5% and those in LSC, SSC, and IR regions were 36.9%, 33.5%, and 42.5%, respectively. A maximum likelihood phylogenomic analysis showed that Z. acanthopodium was evolutionarily closest to Z. piperitum and Z. tragodes ().
Acknowledgments
We thank Dequan Zhang in Dali University for affording plant material of molecular experiments.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data supporting the finding of this study is available in GenBank. The accession number is MT795653: https://www.ncbi.nlm.nih.gov/nuccore/MT795653.
Additional information
Funding
References
- Bhatt V, Kumar N, Sharma U, Singh B. 2018. Comprehensive metabolic profiling of Zanthoxylum armatum and Zanthoxylum acanthopodium leaves, bark, flowers and fruits using ultra high performance liquid chromatography. Sep Sci Plus. 1(5):311–324.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120.
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Editorial Committee of Chinese Journal of Plant of Chinese Academy of Sciences. 1997. Flora of China. Vol. 43. Beijing: Science Press.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Mark-Owitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Majumder M, Sharma HK, Zaman K, Lyngdoh W. 2014. Evaluation of physico-chemical properties and antibacterial activity of the essential oil obtained from the fruits of Zanthoxyllum acanthopodium DC. collected from Meghalaya. Int J Pharm Pharm Sci. 6:543–546.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yang JB, Li DZ, Li HT. 2014. Highly effective sequencing whole chloroplast genomes of angiosperms by nine novel universal primer pairs. Mol Ecol Resour. 14(5):1024–1031.
- Yanti TEP, Nerissa N, Katarina J. 2011. Lemon Pepper Fruit Extract (Zanthoxylum acanthopodium DC.) suppresses the expression of inflammatory mediators in lipopolysaccharide-induced macrophages in vitro. Am J Biochem Biotechnol. 7:190–195.