Abstract
Salix polaris is a dwarf species of the genus Salix which distribute in harsh arctic environment. In this study, we sequenced and assembled whole mitochondrial genome of S. polaris for the first time. The complete mitochondrial genome sequence of this species is a circular molecule of 562920 bp in size, encoding 23 CDS, 21 tRNA genes, and 3 rRNA genes. Mitogenome based phylogenetic tree showed that Salix polaris clustered in the robust clade consist of all Salix species sampled in this study. The complete mitochondrial genome of Salix polaris adds new data into the whole mitochondrial genome of Salix, which is still scarce so far, and we anticipate the accumulation of mitogenome of Salix will facilitate phylogenetic study for this systematic difficult genus.
Salix polaris Wahlenb. is a member of the genus Salix with unique morphological traits, i.e. its a procumbent, dwarf shrub willow plant with typical heights usually less than 9 cm, which distribute in harsh environment of arctic region (Argus Citation1997). This species is an important element of the arctic tundra flora (Skvortsov Citation1999).
In this study, we sequenced and assembled the complete mitochondrial genome of S. polaris. The leaf sample was collected from Svalbard, Norway and dried with silica gel. The voucher specimen was deposit in the Herbarium of Kunming Institute of Botany (accession number: S.G. Wu & al. s.n.) (Chen et al. Citation2010). We used a modified CTAB method (Porebski Citation1997) to isolate the target genomic DNA. The genomic DNA was fragmented and used to construct a short-insert library following the manufacturer’s protocol (Illumina Inc., USA), and then paired-end sequenced on an Illumina Hiseq X Ten sequencer platform. A total of ca. 31.6 million reads generated and assembled by NOVOPlasty v2.7.2 with a k-mer of 39 (Dierckxsens et al. Citation2016). The raw data has been uploaded to NCBI Sequence Read Archive (SRA), the SRA number is SRR:12534577. The complete mitochondrial genome was annotated by Geneious v2020.1.1 software (Kearse et al. Citation2012) using the Salix purpurea as reference.
The mitochondrial genome of S. polaris was assembled as a closed-circular molecule of 5,62,920 b in length (GenBank accession No.: MT806746), containing 23 protein-coding genes (CDS), 21 tRNA genes, and 3 rRNA genes. The overall base composition is 27.4% A, 27.6% T, 22.4% C, 22.7% G, and AT content is higher than GC content. The AT content of protein-coding genes, tRNA genes, rRNA genes was 57.4, 60.2, 60.1%, respectively. All five categories of mitochondrial protein-coding genes that are typically conserved among plants (Chen et al. Citation2017) are present in our assembled genome.
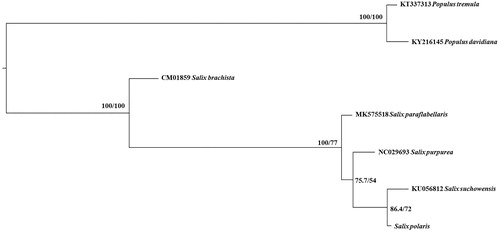
To perform phylogenetic inference based on mitogenome, we download published Salicaceae mitochondrial whole genome (Chen et al. Citation2019; Choi et al. Citation2017; Huang et al. Citation2019; Kersten et al. Citation2016; Wei et al. Citation2016; Ye et al. Citation2017). Maximum-likelihood phylogenetic tree was conducted using IQ_TREE v1.6.10 with 10000 replicates (Nguyen et al. Citation2015) based on 22 mitochondrial coding sequence (CDS) shared by all seven Salicaceae species used in this study which were aligned by software MAFFT v7.47 (Katoh and Standley Citation2013). The best-fitted model is F81 + F + I in terms of Bayesian information criterion using Model Finder (Kalyaanamoorthy et al. Citation2017). We compared the phylogenetic trees of seven species complete mitochondrial genomes of Salicaceae (Chen et al. Citation2019; Huang et al. Citation2019). Our result based on mitogenome phylogenomics showed that all five Salix species and two outgroup Populus species each formed robust monophyletic clade which is sister to each other, confirmed the membership of Salix polaris of the Salix genus and consistent with former phylogeny study of Salicaceae (Chen et al, Citation2010; Huang et al. Citation2017). Salix is a taxa difficult in systematics, we believe that the complete mitochondrial genome of Salix polaris adds new data into the whole mitochondrial genome of Salix, which is still scarce so far, and we anticipate the accumulation of mitogenome of Salix species will facilitate phylogenetic study for this systematic difficult genus.
Disclosure statement
The authors declare that they have no competing interests.
Data availability statement
The data of our research results are available in GenBank at https://www.ncbi.nlm.nih.gov/, Accession number MT806746, SRR12534677.
Additional information
Funding
References
- Argus GW. 1997. Infrageneric classification of Salix (Salicaceae) in the new world. Systemat Bot Monograph. 52:1–121.
- Chen JH, Huang Y, Brachi B, Yun QZ, Zhang W, Lu W, Li HN, Li WQ, Sun XD, Wang GY, et al. 2019. Genome-wide analysis of cushion willow provides insights into alpine plant divergence in a biodiversity hotspot. Nat Commun. 10(1):5230.
- Chen JH, Sun H, Wen J, Yang YP. 2010. Molecular phylogeny of Salix L.(Salicaceae) inferred from three chloroplast datasets and its systematic implications. Taxon. 59(1):29–37.
- Chen ZW, Zhao N, Li SS, Grover CE, Nie HS, Wendel JF, Hua JP. 2017. Plant mitochondrial genome evolution and cytoplasmic male sterility. Crit Rev Plant Sci. 36(1):55–69.
- Choi MN, Han M, Lee H, Park HS, Kim MY, Kim JS, Na YJ, Sim SW, Park EJ. 2017. The complete mitochondrial genome sequence of Populus davidiana dode. Mitochondr DNA B. 2(1):113–114.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Huang Y, Chen X, Chen KY, Li WQ, Shi MM, Chen JH. 2019. The complete mitochondrial genome of Salix paraflabellaris, an endemic alpine plant of Yunnan province of China. Mitochondrial DNA Part B. 4(1):1394–1395.
- Huang Y, Wang J, Yang YP, Fan CZ, Chen JH. 2017. Phylogenomic analysis and dynamic evolution of chloroplast genomes in Salicaceae. Front Plant Sci. 8:1050.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. Mafft multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kersten B, Rampant PF, Mader M, Paslier ML, Bounon R, Berard A, Vettori C, Schroeder H, Leple J, Fladung M. 2016. Genome sequences of Populus tremula chloroplast and mitochondrion: implications for holistic poplar breeding. PLOS One. 11(1):e0147209.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15.
- Skvortsov AK. 1999. Willows of Russia and adjacent countries: Taxonomical and geographical revision. Joensuu: University of Joensuu.
- Wei S, Wang X, Bi C, Xu Y, Wu D, Ye N. 2016. Assembly and analysis of the complete Salix purpurea L. (Salicaceae) mitochondrial genome sequence. Springerplus. 5(1):1894–1894.
- Ye N, Wang X, Li J, Bi C, Xu Y, Wu D, Ye Q. 2017. Assembly and comparative analysis of complete mitochondrial genome sequence of an economic plant Salix suchowensis. PeerJ. 5:e3148.