Abstract
We report the first complete mitogenome (Mt) sequence of Anopheles coustani, an understudied malaria vector in Africa. The sequence was extracted from one individual mosquito from São Tomé island. The length of the A. coustani Mt genome was 15,408 bp with 79.3% AT content. Phylogenetic analysis revealed that A. coustani is most closely related to A. sinensis (93.5% of identity); and 90.1% identical to A. gambiae complex members.
Anopheles coustani was first described by Laveran (1900) from specimens collected in Madagascar. This species is widespread and abundant over much of the African continent (Coetzee Citation1994). However, this species has been understudied because it is widely considered as a secondary malaria vector. Recent studies have questioned its supposed negligible importance in malaria transmission, since specimens of A. coustani were persistently found infected with Plasmodium in Cameroon (Antonio-Nkondjio et al. Citation2006), Kenya (Mwangangi et al. Citation2013) and Madagascar (Nepomichene et al. Citation2015; Tedrow et al. Citation2019). Here, we report the complete mitogenome (Mt) sequence of a field-collected A. coustani from São Tomé island (O°22’N 6°42’E). The specimen sequenced was an adult female collected by human landing catch outside of houses in December 2019. Anopheles coustani and A. gambiae s.l are the only anophelines and potential malaria vectors found on the island (Pinto et al. Citation2000).
DNA was extracted using a Qiagen Biosprint following our established protocol (Nieman et al. Citation2015). Library preparation was conducted using 10 ng of genomic DNA as input, as described by Yamasaki et al. (Citation2016). Size selection of the library and clean-up was performed using AMPure SPRI beads (Beckman Coulter Life Sciences, Indianapolis, Indiana, USA). The library was sequenced for 150 bp paired-end reads using a HiSeq 4000 instrument (Illumina, San Diego, CA) at Novogene Corporation. Raw-sequencing reads were used to assemble the Mt contig using NOVOPlasty version 2.6.7 (Dierckxsens et al. Citation2017).
The length of the A. coustani Mt (Genbank: MT806097) is 15,408 bp and the percentage of A + T was 79.3%. The 672 bp long COI fragment spanning 1464-2135 of the A. coustani Mt is highly similar to the COI fragment sequences of A. coustani from Mali (Huestis et al. Citation2019), Guinea-Bissau (Gordicho et al. Citation2014) and Kenya (St Laurent et al. Citation2016) with an average pairwise identity of 99.4% (±0.2 SD). The 622 bp long COII fragment spanning Mt:3054-3675 was 99.7% identical to the published A. coustani COII sequence from Gabon (Ayala et al. Citation2019). In addition, we sequenced the internal transcribed spacer 2 (ITS2; GenBank: MT791041) region of nuclear ribosomal DNA which was 99.64% identical to the published A. coustani from Guinea (Cansado-Utrilla et al. Citation2020).
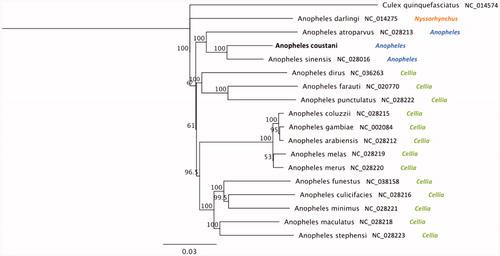
A phylogeny including other primary and secondary malaria vector species and Culex quinquefasciatus as an outgroup is illustrated in . The Jukes-Cantor model was used to calculate pairwise genetic distances and the neighbour-joining method was used to build the phylogenetic tree using Geneious Prime 2020.1.2 (https://www.geneious.com). Anopheles coustani was most closely related to the Asian malaria vector, Anopheles sinensis (93.5% of identity); and 90.1% on average sequence similarity with African malaria vectors of the A. gambiae complex (A. gambiae s.s., A. coluzzii, A. arabiensis, A. melas, A. merus). These results are consistent with the formal taxonomic arrangement of these species in which A. coustani and A. sinensis are in the same subgenus, Anopheles, whereas A. gambiae s.l. is in the subgenus Cellia (Knight and Stone Citation1977). The extracted DNA sample is maintained in the Vector Genetics Laboratory specimen archive at UC Davis with Accession ID 1219-ST-PGA3-033 and metadata record is available through https://popi.ucdavis.edu/.
Figure 1. Phylogenetic tree based on mitogenome sequences of anophelines. GenBank IDs and anopheline’ subgenus are provided next to each species name. Numbers at nodes indicate bootstrap values out of 200 replicates. Culex quinquefasciatus was considered as an outgroup. Branch length scale bar indicates relative differences (0.03 = 3% nucleotide difference).
The mitogenome was annotated using MITOS (Bernt et al. Citation2013) using the invertebrate genetic code under default settings. The gene start and end positions and gene orders were by and large consistent with published annotations of mosquito Mt sequences used in the phylogeny. This new annotated mitogenome, from which additional genetic markers may be retrieved, will serve as a basis for subsequent mtDNA-based phylogenetic analyses of this species.
Acknowledgements
We thank the University of California-Irvine, Malaria Initiative (UCIMI) and The Open Philanthropy Project for their support. We thank the National Malaria Control Program personnel and, Ministry of Health in São Tomé and Príncipe who facilitated our field collections in São Tomé. We thank Dr. Travis C. Collier (UC Davis) who has maintained the genomics data analysis throughout this project.
Disclosure statement
No potential conflict of interest was report by the author(s).
Data availability statement
The data that support the findings of this study are available in Genbank (mitogenome: MT806097; ITS2: MT791041). The meta data associated with it as well as the access to the DNA specimen is available through PopI (https://popi.ucdavis.edu/) with sample ID 1219-ST-PGA3-033.
Additional information
Funding
References
- Antonio-Nkondjio C, Kerah CH, Simard F, Awono-Ambene P, Chouaibou M, Tchuinkam T, Fontenille D. 2006. Complexity of the malaria vectorial system in Cameroon: contribution of secondary vectors to malaria transmission. J Med Entomol. 43(6):1215–1221.
- Ayala D, Akone-Ella O, Rahola N, Kengne P, Ngangue MF, Mezeme F, Makanga BK, Nigg M, Costantini C, Simard F, et al. 2019. Natural Wolbachia infections are common in the major malaria vectors in Central Africa. Evol Appl. 12(8):1583–1594.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Cansado-Utrilla C, Jeffries CL, Kristan M, Brugman VA, Heard P, Camara G, Sylla M, Beavogui AH, Messenger LA, Irish SR, et al. 2020. An assessment of adult mosquito collection techniques for studying species abundance and diversity in Maferinyah, Guinea. Parasit Vectors. 13(1):150.
- Coetzee M. 1994. Anopheles crypticus, new species from South Africa is distinguished from Anopheles coustani. Mosquito Systematics. 26:125–131.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Gordicho V, Vicente JL, Sousa CA, Caputo B, Pombi M, Dinis J, Seixas G, Palsson K, Weetman D, Rodrigues A, et al. 2014. First report of an exophilic Anopheles arabiensis population in Bissau City, Guinea-Bissau: recent introduction or sampling bias? Malar J. 13(1):423.
- Huestis DL, Dao A, Diallo M, Sanogo ZL, Samake D, Yaro AS, Ousman Y, Linton YM, Krishna A, Veru L, et al. 2019. Windborne long-distance migration of malaria mosquitoes in the Sahel. Nature. 574(7778):404–408.
- Knight KL, Stone A. 1977. A catalog of the mosquitoes of the world. Lanham: Entomological Society of America.
- Mwangangi JM, Muturi EJ, Muriu SM, Nzovu J, Midega JT, Mbogo C. 2013. The role of Anopheles arabiensis and Anopheles coustani in indoor and outdoor malaria transmission in Taveta District, Kenya. Parasit Vectors. 6:114.
- Nepomichene TN, Tata E, Boyer S. 2015. Malaria case in Madagascar, probable implication of a new vector, Anopheles coustani. Malar J. 14:475.
- Nieman CC, Yamasaki Y, Collier TC, Lee Y. 2015. A DNA extraction protocol for improved DNA yield from individual mosquitoes. F1000Res. 4:1314.
- Pinto J, Sousa CA, Gil V, Ferreira C, Gonçalves L, Lopes D, Petrarca V, Charlwood JD, Rosário VE. 2000. Malaria in São Tomé and Príncipe: parasite prevalences and vector densities. Acta Trop. 76(2):185–193.
- St Laurent B, Cooke M, Krishnankutty SM, Asih P, Mueller JD, Kahindi S, Ayoma E, Oriango RM, Thumloup J, Drakeley C, et al. 2016. Molecular characterization reveals diverse and unknown malaria vectors in the western Kenyan highlands. Am J Trop Med Hyg. 94(2):327–335.
- Tedrow RE, Rakotomanga T, Nepomichene T, Howes RE, Ratovonjato J, Ratsimbasoa AC, Svenson GJ, Zimmerman PA. 2019. Anopheles mosquito surveillance in Madagascar reveals multiple blood feeding behavior and Plasmodium infection. PLoS Negl Trop Dis. 13(7):e0007176.
- Yamasaki YK, Nieman CC, Chang AN, Collier TC, Main BJ, Lee Y. 2016. Improved tools for genomic DNA library construction of small insects. [version 1; not peer reviewed]. F1000Research 2016, 5:211.https://doi.org/10.7490/f1000research.1111322.1
