Abstract
Rhinogobius duospilus is a small freshwater fish with brilliant color in southern China, belonging to the subfamily Gobionellinae. In this study, the complete mitochondrial genome of 16,496 bp from R. duospilus was reported for the first time. It composed of 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and 2 non-coding genes. Phylogenetic tree showed that R. duospilus formed a separate lineage. The findings here would be helpful to further researches of R. duospilus.
Rhinogobius duospilus is a small benthic freshwater fish in the subfamily Gobionellinae (Teleostei: Gobiidae) that is mainly distributed in southern China and classified as a new species since 1935 (Herre Citation1935, Li et al. Citation2008). Because of the unique spawning behavior and colorful appearance, it is well-liked by ornamental fish lovers. Previous studies of R. duospilus had focused on taxonomy and morphology, and the population structure based on genetic markers had been reported until recently (Feng et al. Citation2017; Li et al. Citation2008; Wu et al. Citation2016), but the characterization of the complete mitogenome remain unknown. Here, we reported the complete mitochondrial genome of R. duospilus for the first time, contributing to the phylogenetic and genetic protection researches of this species.
Specimens of R. duospilus were collected from Guilin City, Guangxi, China (25°16′25.00″N, 100°17′24.07″E) and were stored at −20 °C in Guangxi Colleges and Universities Key Laboratory of Aquatic Healthy Breeding and Nutrition Regulation, Guangxi University. The total genomic DNA was extracted from the dorsal muscle. The sequences were amplified by PCR with twenty pairs of primers and then the mitochondrial genome was annotated though MitoAnnotator (Iwasaki et al. Citation2013).
The complete mitochondrial genome of R. duospilus was 16,496 bp in length (GenBank accession no. MH127918), with A + T-biased (the G + C content was 47.3%). The overall base nucleotide composition was 27.1% A, 30.3% C, 17.0% G, 25.7% T. The mitochondrial genome contains 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, 2 non-coding genes (light strand origin of replication (OL) and control region (D-loop)), which were high degree of conservation among R. duospilus and other Gobionellinae fishes. The 12S and 16S rRNA genes were 953 and 1684 bp in length, respectively. The OL was 32 bp in length and located between tRNAAsn and tRNACys and the D-loop was 843 bp in length and located between tRNAPro and tRNAPhe. Among the 13 PCGS, there were 12 genes with ATG as the start codon, except for COI with GTG as the start codon. In addition, six PCGS had a complete TAA stop codon and two PCGs had a complete TAG stop codon, while five PCGs (COII, COIII, ND3, ND4 and CYTB) showed an incomplete stop codon (T).
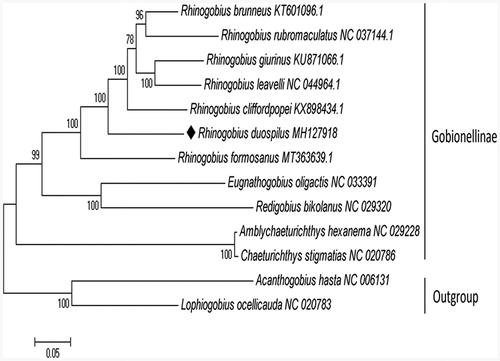
The maximum likelihood tree was constructed based on 13 mitochondrial PCGs of 11 Gobionellinae species using MEGA 7.0 software (Kumar et al. Citation2016). Acanthogobius hasta and Lophiogobius ocellicauda were included as outgroups. Evolutionary model was inferred by the program PhyML 3.0, where the substitution model GTR + G+I was considered as the most appropriate to the data (Guindon et al. Citation2010). Compared with the other Gobionellinae species, R. duospilus formed a separate lineage, which was showed in the phylogenetic tree (). These results provided an important basis for further studies on mitochondrial genome and phylogenetics of R. duospilus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/nuccore/MH127918, GenBank Accession Number: MH127918.
Additional information
Funding
References
- Feng WL, Wu ZQ, Huang LL, Hu YX, Shi RD, Ding Y, Chang XZ. 2017. Morphological characters of 16 larval and juvenile fish species in the middle reaches of Lijiang river. J Hydr. 38:1674–3075.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Herre AWCT. 1935. Note on the fishes in the Zoological Museum of Stanford University. VI. New and rare Hong Kong fishes obtained in 1934. Hong Kong Naturalist. 6:285–293.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. Mitofish and Mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Li L, Fan L, Zhong JS. 2008. Study on the early development of Rhinogobius duospilus larvae. J Shanghai Fish Univ. 17:447–451.
- Wu TH, Tsang LM, Chen IS, Chu KH. 2016. Multilocus approach reveals cryptic lineages in the goby Rhinogobius duospilus in Hong Kong streams: Role of paleodrainage systems in shaping marked population differentiation in a city. Mol Phylogenet E. 104:112–122.