Abstract
The mitogenome of Phyllidiopsis krempfi has been determined for the first time. The assembled mitogenome was 14,970 bp in length, including 13 protein-coding genes, 22 transfer RNA, and 2 ribosomal RNA genes. The gene content and order were identical with those of the other Phyllidiid species. The molecular taxonomic position of P. krempfi was clustered with the Phyllidiid species. The genus Phyllodiopsis clade is closely related with the genus Phyllidia. The mitogenome of P. krempfi provides significant DNA molecular data for further identification and phylogenetic analysis within the Phyllidiid.
Nudibranchs are called ‘ocean cleaners’ likened to hyenas on land because they feed on sponges, hydoids (Folino Citation1997), bryozoans (Dominguez et al. Citation2008), etc. The genus Phyllidiopsis Bergh, Citation1876 including the Phyllidiidae are distributed mostly throughout Indo-Pacific tropical waters, which comprises 37 valid species (Valdés Citation2001; MolluscaBase Citation2020). Its members are characterized by having an elongate foregut and fused oral tentacles (Brunckhorst Citation1993). The phylogenetic relationships within the Phyllidiidae based on molecular and morphology analyses has been conducted, but position of the genera within the family remains unclear (Brunckhorst Citation1993; Valdés Citation2002; Valdés Citation2003; Stoffels Citation2016). The mitochondrial genome is a useful tool for inferring phylogenetic relationships (Matsudaira and Ishida Citation2010). In this study, we analyzed the full-length mitochondrial genome (mitogenome) of the P. krempfi for the first time and it is valuable information for further study on molecular systematics and phylogenetic relationships within the Phyllidiidae.
Specimens of P. krempfi were collected from the Pacific Ocean (7°16′20.02′N, 134°31′21.68′E). The voucher specimens are deposited in the National Marine Biodiversity Institute of Korea (MABIK Lot no. 0018233). The genomic DNA was extracted from the muscle tissue and the mitogenome sequences were analyzed in two ways: first, after the COI gene was amplified through the universal primers, the primer sets were designed from the partial sequences of the COI gene of P. krempfi and highly preservative gene regions for the Phyllidiopsis species. Then, we conducted the long-range PCR (LR-PCR) to amplify the targeted genomic intervals and sequenced by the Sanger method. The sequences were assembled and annotated in comparison with the previously reported mitogenome sequences of the Phyllidiid species (Yu et al. Citation2018; Dinh Do et al. Citation2019) using the Geneious v9.1.2 (Kearse et al. Citation2012). Additionally, the mitochondrial genome annotation (MITOS) server (Bernt et al. Citation2013), and the tRNAscan-SE server (Lowe and Chan Citation2016) were used for annotation.
The assembled P. krempfi mitogenome (GenBank accession number MT726194) was a 14,970 bp long circular DNA with 2 ribosomal RNA (rRNA), 22 transfer RNA (tRNA), and 13 protein-coding genes (PCGs). The gene content and order were identical to those of the species belonging to the order Nudibranchia (Xiang et al. Citation2016; Yu et al. Citation2018; Dinh Do et al. Citation2019). All PCGs get off by the typical ATG as start codon. Eight (COI, nad6, nad1, nad4l, CO2, atp6, nad3, nad4) of 13 PCGs use TAA for the stop codon, and three genes (nad5, atp8, CO3) ends with TAG while nad2 and cytb genes have an incomplete stop codon, T.
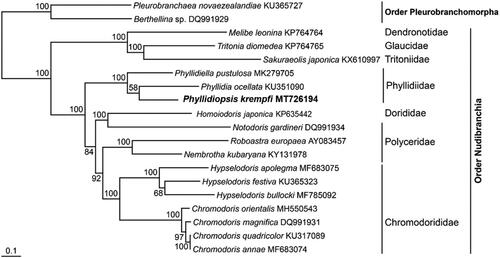
A maximum-likelihood (ML) tree was constructed to investigate the molecular taxonomic position of these species using RAxML 8.2 (Stamatakis Citation2014), and the dataset used the nucleotide sequences of the 12 PCGs excluding atp8 of the other species belonging to the Nudibranchia. Phyllidiopsis krempfi was clustered with the Phyllidiid species previously announced from the GenBank, with high bootstrap values of 100% (). The genus Phyllodiopsis clade is closely related with the genus Phyllidia, as well as support for the previously published 16s gene tree (Valdés Citation2003).
Figure 1. Maximum-likelihood (ML) phylogeny based on the full-length mitochondrial genomes from the marine invertebrate species belonging to the order Nudibranchia. The nucleotide sequence matrix included the first, second and third codon positions of the 12 protein-coding genes. A bootstrap value above 50% in the ML analysis is indicated at each node. Phyllidiopsis krempfi analyzed in this study is shown in bold.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT726194.
Additional information
Funding
References
- Bergh R. 1876. Neue Beiträge zur Kenntniss der Phyllidiaden. Verhandlungen der Königlich-Kaiserlich Zoologisch-Botanischen Gesellschaft in Wien. Abhandlungen. 25:659–674.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Brunckhorst DJ. 1993. The systematics and phylogeny of phyllidiid nudibranchs (Doridoidea). Rec Aust Mus, Suppl. 16:1–18.
- Dinh Do T, Choi TJ, Jung DW, Kim JI, Karagozlu MZ, Kim CB. 2019. The complete mitochondrial genome of Phyllidiella pustulosa (Cuvier, 1804) (Nudibranchia, Phyllidiidae). Mitochondrial DNA Part B. 4(1):771–772.
- Dominguez M, Troncoso JS, Garcia FJ. 2008. The family Aeolidiidae Gray, 1827 (Gastropoda Opisthobranchia) from Brazil, with a description of a new species belonging to the genus Berghia Trinchese, 1877. Zool J Linn Soc. 153(2):349–368.
- Folino NC. 1997. The role of prey mobility in the population ecology of the nudibranch Cuthona nana (Gastropoda: Opisthobranchia). Amn Malacol Bull. 14:17–26.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–57.
- Matsudaira K, Ishida T. 2010. Phylogenetic relationships and divergence dates of the whole mitochondrial genome sequences among three gibbon genera. Mol Phylogenet Evol. 55(2):454–459.
- MolluscaBase. 2020. Phyllidiopsis krempfi Pruvot-Fol, 1957. World Register of Marine Species [cited 30 Jun 2020]. https://www.marinespecies.org/aphia.php?p=taxdetails&id=536716on
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Stoffels BE, van der Meij SE, Hoeksema BW, van Alphen J, van Alen T, Meyers-Muñoz MA, de Voogd NJ, Tuti Y, van der Velde G. 2016. Phylogenetic relationships within the Phyllidiidae (Opisthobranchia, Nudibranchia). ZK. 605:1–35.
- Valdés A. 2001. Depth-related adaptations, speciation processes and evolution of color in the genus Phyllidiopsis (Mollusca: Nudibranchia). Mar Biol. 139(3):485–496. cryptobranch dorid
- Valdés A. 2002. A phylogenetic analysis and systematic revision of the s (Mollusca, Nudibranchia, Anthobranchia). Zool J Linnean Soc. 136(4):535–636.
- Valdés A. 2003. Preliminary molecular phylogeny of the radula-less dorids (Gastropoda: Opisthobranchia), based on 16S mtDNA sequence data. J Molluscan Stud. 69(1):75–80.
- Xiang P, Lin M, Wang Y, Shen KN, Hsiao CD. 2016. The complete mitogenome of sea slug, Phyllidia ocellata (Mollusca: Phyllidiidae). Mitochondrial DNA Part B. 1(1):96–97.
- Yu C, Kim H, Kim HJ, Jung YH. 2018. The complete mitochondrial genome of the Oriental sea slug: Chromodoris orientalis (Nudibranchia, Chromodorididae). Mitochondrial DNA Part B. 3(2):1017–1018.
