Abstract
Acanthopanax trifoliatus (Linn.) Merr. is an edible vegetables and medicinal plant from Asian countries. In this study, the complete chloroplast genome of A. trifoliatus was assembled and annotated by high-throughput sequencing. The total chloroplast genome size of A. trifoliatus was 156,716 bp, containing a large single-copy (LSC) region of 86,672 bp, a small single-copy (SSC) region of 18,174 bp, and a pair of inverted repeat regions of 25,935 bp. A total of 134 genes were predicted in the chloroplast genome of A. trifoliatus, including 89 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that A. trifoliatus was closely related to Eleutherococcus gracilistylus.
Acanthopanax trifoliatus is an edible medicinal plant belonging to the Acanthopanax genus in the Araliaceae family. It is commonly cultivated in China, India, Thailand, Vietnam, and the Philippines. Leaves of A. trifoliatus are popularly consumed as vegetables and herbal tea in southern China (Chen et al. Citation2019). Traditionally, A. trifoliatus has also been used as folk medicine to treat various diseases, including lung hemorrhages, bruises, ulcers, and partial (Sithisarn et al. Citation2009). Pharmacological studies also reported that A. trifoliatus extracts showed numerous biological activities, including antioxidative, anti-inflammatory, anti-cancer, and neuroprotective effects (Chen et al. Citation2019). The sequencing of the chloroplast genome of medicinal plants will promote in-depth research on the chloroplast genome of medicinal plants and the development of related industries. In this study, we assembled the complete chloroplast genome of A. trifoliatus to provide genomic and genetic sources for further research.
The fresh leaves of A. trifoliatus were collected from Guangdong Academy of Agricultural Sciences (Guangzhou, China, N23.1459, E113.3498). The samples were frozen in liquid nitrogen and stored at −80 °C refrigerator until use at Key Laboratory for Crops Genetic Improvement of Guangdong in Guangdong Academy of Agricultural Sciences (specimen code LC2020). Total genomic DNA was extracted using plant genomic DNA kit (Omega, Norcross, GA) and then constructed DNA library with average insert size 350-bp using VAHTS Universal DNA Library Prep Kit (Vazyme ND606-01, Nanjing, China). Finally, the library was sequenced on the Novaseq platform (Illumina, San Diego, CA) following as recommended by the manufacturer. The sequence was assembled by the GetOrganelle (Jin et al. Citation2019) and annotated with the Geseq (Tillich et al. Citation2017) using the cp annotation of Fatsia japonica (NC_027685) as a reference. Then, the chloroplast genome sequence was submitted to GenBank with the accession number MT754220.
The total genome size of A. trifoliatus was 1,56,716 bp, containing an large single-copy (LSC) region of 86,672 bp, a small single-copy (SSC) region of 18,174 bp, and a pair of IRs region of 25,935 bp. A total of 134 genes were predicted, including 89 protein-coding genes, 37 tRNA genes, and eight rRNA genes, and the GC content was 38.02%.
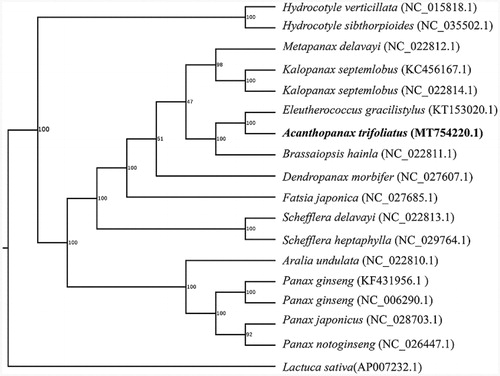
Phylogenetic analysis suggested that A. trifoliatus was closely clustered with Eleutherococcus gracilistylus (), which was generated based on the 18 complete chloroplast genomes using maximum likelihood method by RaxML software (Stamatakis Citation2014) using the GTRGAMMA nucleotide substitution model. The chloroplast genome sequence of A. trifoliatus will provide useful genetic information for further study on molecular breeding and genetic engineering.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank (https://www.ncbi.nlm.nih.gov/nuccore/ MT754220.1), and the assession number is MT754220.1.
Additional information
Funding
References
- Chen M, Qin Y, Ma H, Zheng X, Zhou R, Sun S, Huang Y, Duan Q, Liu W, Wu P, et al. 2019. Downregulating NF-κB signaling pathway with triterpenoids for attenuating inflammation: in vitro and in vivo studies. Food Funct. 10(8):5080–5090.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. 256479.
- Sithisarn P, Jarikasem S, Thisayakorn K. 2009. Anti-inflammatory and antioxidative effects of leaf extract from Acanthopanax trifoliatus. Planta Med. 75(9):891–891.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.