ABSTACT
An important horticultural moth orchid in China, Phalaenopsis lobbii, has small and lovely flowers with unique labial characteristics. We sequenced the chloroplast genome (cpDNAs) of P. lobbii and analyzed the structural characteristics of its complete cpDNAs sequence. The results showed that P. lobbii had a typical circular double-stranded tetrad structure with the average GC content of 36.60%, and its total length was 144,607 bp. The complete chloroplast was annotated with 125 genes, eight rRNAs and 38 tRNAs. Phylogenetic tree displayed that P. lobbii was closely related to P. equestris, P. aphrodite subsp. formosana, P. ‘Tiny Star’ and P. japonica. This published cpDNAs of P. lobbii will provide information for future phylogenetic and artificial breeding of Phalaenopsis.
Phalaenopsis (Orchidaceae), such as butterflies, is enchanting and charming, with a broad market prospect (Zhu Citation2004). It is divided into five subgenera, subgen. Proboscidioides, subgen. Aphyllae, subgen. Parishianae, subgen. Polychilos and subgen. Phalaenopsis (Christenson Citation2001). Phalaenopsis lobbii is a member of subgen. Parishianae, named in honor of Thomas Lobb, an English plant collector. Unlike other phalaenopsis, P. lobbii has a light yellow labellum with a white line in the center, and its pendulous is inverted triangle (Chen and Ji Citation2000; Huang et al. Citation2020). It has important breeding value because its unique labial features can be passed on to offspring and produce special patterns. However, the genetic information related to P. lobbii is not yet clear, and the phylogeny of subgen. Parishianae is controversial (Tsai et al. Citation2010). The publication of the chloroplast genome (cpDNAs) sequence of P. lobbii will help to further clarify the origin and evolution of Phalaenopsis and promote the development of new cultivars.
The cultivated plants of P. lobbii were sampled from Yunnan, China, and deposited in Fujian Agriculture and Forestry University (FAFU). The specimens for sequencing were stored in the laboratory of FAFU (Voucher specimen: HDL-FJ2019-15A, 26°20′21.3″N, 113°12′39.6″E). We used a modified CTAB-based method to extract the DNA of the fresh young leaves of its plant, used the BGISEQ-500 platform (BGI, Wuhan, China) for sequencing (Zhou et al. Citation2019). Fastp software removed automatically low-quality reads and adapters, resulting in approximately 15 Gb of clean reads (Chen et al. Citation2018). Then, the cpDNAs sequence assembly was conducted using SPAdes version 3.13.1 software, formed a circular chloroplast genome. Taking the notes of P. equestris (NC_017609.1) as a reference, the MAFFT plugin in Geneious version 2020.2.2 was used for contrast annotation, and manual adjustment was conducted in combination with the annotation data of Organellar Genome DRAW (OGDRAW) (Stephan et al. Citation2019). Finally, the size of the complete cpDNAs sequence of P. lobii (GenBank accession MT830847) is 144,607 bp, including an LSC region (83,482 bp), an SSC region (11,687 bp), and two IRs regions (24,719 bp). The average GC content was 36.6%, and the complete chloroplast contains 125 genes, eight rRNAs and 38 tRNAs.
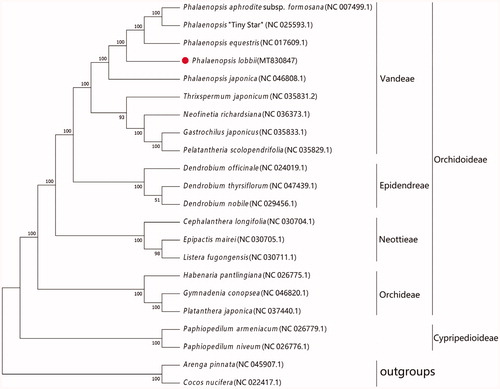
Using Arenga pinnata and Cocos nucifera as outgroups, P. lobbii and 19 published complete cpDNAs of Orchidaceae construct a maximum likelihood (ML) phylogeny by MAFFT version 7 (Katoh et al. Citation2019) and MEGA version 7 (Kumar et al. Citation2016) (). The phylogenetic tree showed that P. lobbii and its sisters (P. equestris, P. aphrodite subsp. formosana, P. ‘Tiny Star’ and P. japonica) constituted a single clade. The complete cpDNAs of P. lobbii will provide important information for studies on the origin and evolution of Phalaenopsis and breeding techniques.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The Phalaenopsis lobbii raw data has been stored in omics database of Genome Sequence Archive. GSA accession number is CRA003144. All the information can be found on the website (https://bigd.big.ac.cn/gsa/browse/CRA003144/CRX150835).
Additional information
Funding
References
- Chen XQ, Ji ZH. 2000. Chinese orchids. Beijing: China Forest Industry press.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–890.
- Christenson EA. 2001. Phalaenopsis: a monograph. USA: Timber Press Portland.
- Stephan G, Pascal L, Ralph B. 2019. Organellar Genome DRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(1):59–64.
- Huang XY, Xie ZX, Lu ZZ, et al. 2020. Sterile seeding and rapid propagation of Phalaenopsis lobbii. Plant Physiol J. 56(4):693–699.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinformatics. 20(4):1160–1166.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Tsai CC, Chiang YC, Huang SC, Chen CH, Chou CH. 2010. Molecular phylogeny of Phalaenopsis Blume (Orchidaceae) on the basis of plastid and nuclear DNA. Plant Syst Evol. 288(1–2):77–98.
- Zhou Y, Liu C, Zhou R, Lu A, Huang B, Liu L, Chen L, Luo B, Huang J, Tian Z, et al. 2019. SEQdata-BEACON: a comprehensive database of sequencing performance and statistical tools for performance evaluation and yield simulation in BGISEQ-500. BioData Mining. 12(1):1.
- Zhu GF. 2004. Phalaenopsis. Guangzhou: Guangdong Science and Technology Press.