Abstract
The complete chloroplast genome sequence of Astilboides tabularis, one of endemic species of Eastern Asia, was determined. The chloroplast genome was 157,147 bp in length with large single-copy (87,703 bp), small single-copy (18,268 bp) and a pair of inverted repeats (25,588 bp). In total, 131 genes were encoded, including 86 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The phylogenetic analysis using concatenated 77 protein-coding genes of 15 species chloroplast genome revealed that A. tabularis was sister to the clade containing Bergenia, Oresitrophe, and Mukdenia.
Astiboides tubularis (Hemsley) Engler (Saxifragaceae) is the only member of the genus Astiboides and is endemic to North East Asia. This monotypic genus is similar with Rodgersia A. Gray, but different by having large simple leaf and panicle inflorescence (Jintang and Cullen Citation2001; Lee Citation2003). This species is distributed in the regions of north eastern China (i.e. Jilin and Liaoning) and the Korean Peninsula (Lee Citation1996; Jintang and Cullen Citation2001; Yoon et al. Citation2015). In South Korea, A. tabularis was designated as an endangered and rare species and protected by multiple laws, such as Wildlife Protection and Management Act by the Ministry of Environment and Act on the Creation and Furtherance of Arboretums and Gardens by the Korea Forest Service (Lee Citation2009; Kim Citation2014). Furthermore, A. tabularis is an economically important plant resource as an edible and medicinal plant (Chang et al. Citation2005; Yang et al. Citation2020). In addition, its gigantic leaf and inflorescence have horticultural and ornamental values (Cho et al. Citation2016). Therefore, it is important to examine its genetic diversity and structure to conserve genetic resources of this species. As a first attempt, we report complete chloroplast (CP) genome sequence of A. tabularis.
Fresh leaves of A. tabularis were sampled from the living collection (KNKB 2020-15) of Korea National Arboretum, originally collected from Gangwon-do Province (37°29′ N 128°38′ E). Total genomic DNA was isolated using the DNeasy Plant Mini Kit followed by the manufacturer’s protocol (Qiagen Inc., Valencia, CA). Extracted high-quality gDNA was sequenced by using Illumina MiSeq (Illumina Inc., San Diego, CA) platform with 550 bp insert size. Paired-end reads were trimmed in Geneious R v. 10.2.6 program (Biomatters Ltd., Auckland, New Zealand). To construct the CP genome of A. tabularis, Bergenia scopulosa (NC_036061) was used as a reference. The chloroplast genome of A. tabularis was annotated by using the Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005). Phylogenetic tree including 13 species of Saxifragaceae was constructed based on concatenated 77 chloroplast protein coding genes using IQ-TREE v.1.6.8 (Nguyen et al. Citation2015) with 1000 bootstrap replications ().
Figure 1. Maximum-likelihood tree based on 77 protein-coding genes from 13 representative species of Saxifragaceae. Bootstrap support values are shown at the branches.
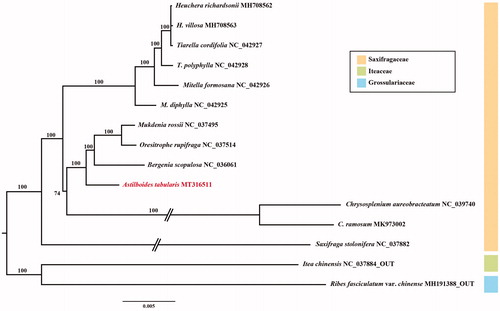
The CP genome of A. tabularis (MT316511) has a typical quadripartite structure (157,147 bp) including large single copy (LSC; 87,703 bp), small single copy (SSC; 18,268 bp) and a pair of inverted repeats (IRa and IRb; 25,588 bp). The chloroplast genome contained 131 genes including 86 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The phylogenetic analysis of Saxifragaceae showed that A. tabularis is sister with the clade containing Bergenia, Oresitrophe, and Mukdenia (bootstrap value = 100%). Further work on details is necessary to elucidate phylogenetic relationships of the monotypic genus, Astilboides, and other major lineages of Saxifragaceae.
Acknowledgements
We are grateful to several researcher and lab associates for assistance in field collection.
Disclosure statement
The authors declare no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT316511.
Additional information
Funding
References
- Chang C-S, Lee H-S, Park T-Y, Kim H. 2005. Reconsideration of rare and endangered plant species in Korea based on the IUCN Red List Categories. Korean J Ecol. 28(5):305–320.
- Cho JS, Jeong JH, Lee CH. 2016. The effects of environmental conditions and chemical treatments on seed germination in Astilboides tabularis (Hemsl.) Engl. Korean J Hortic Sci Technol. 34(3):363–371.
- Jintang P, Cullen J. 2001. Astilboides. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 8. Brassicaceae through Saxifragaceae. Beijing (China)/St. Louis (MI): Science Press/Missouri Botanical Garden Press.
- Kim S-B. 2014. Korean red list of threatened species. 2nd ed. National Institute of Biological Resources.
- Lee B-C. 2009. Rare plants data book in Korea. Pocheon (Korea): Korea National Arboretum.
- Lee TB. 2003. Colored flora of Korea. Seoul (Korea): Hyangmunsa.
- Lee WT. 1996. Lineamenta florae Koreae. Seoul (Korea): Academy Publ.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yang J, Chanok L, Heekyu K, Kwon YS, Kim MJ. 2020. Biological activities and phenolic compound content of Astilboides tabularis (Hemsl.). Engler extracts. Curr Pharm Biotechnol. 21(11):1070–1078.
- Yoon YS, Kim K-A, Yoo K-O. 2015. Environmental characteristics of Astilboides tabularis (Hemsl.) Engl. habitats. Korean J Plant Resour. 28(1):64–78.
