Abstract
Blossom-headed Parakeet Psittacula roseate is listed as Near Threatened on the IUCN Red List of Threatened Species because of its habitat loss, and declining population. In this study, we first sequenced and described the complete mitochondrial genome and phylogeny of P. roseata. The whole genome of P. roseata was 16,814 bp in length, and contained 14 protein-coding genes, 22 transfer RNA genes, 2 ribosome RNA genes, and 1 non-coding control regions. The overall base composition of the mitochondrial DNA was 31.88% for A, 22.00% for T, 32.88% for C, 13.23% for G, with a GC content of 46.11%. A phylogenetic tree strongly supported that genus Psittacula closely related with genus Eclectus by highly probability.
Blossom-headed Parakeet (Psittacula roseate) occurs in the lowlands to c.1,500 m, inhabiting light forest, including savanna, secondary growth, forest edge, clearings and cultivated areas (Juniper and Parr Citation1998). Psittacula roseata occurs in South and South-East Asia, ranging from eastern India and Bangladesh, through Myanmar, Thailand, Laos, Cambodia, Vietnam and southern China (Yunnan, Guangxi and Guangdong provinces) (Juniper and Parr Citation1998). It is particularly associated with dry deciduous forest throughout south-east Asia. It is listed as Near Threatened on the basis that it is undergoing a moderately rapid decline owing to habitat loss and trapping pressure (BirdLife International Citation2019; IUCN. Citation2019). Molecular studies supported closely related with P. roseata and P. cyanocephala by highly probability based on cytochrome-b gene (Kundu et al. Citation2012). Psittacula roseata was the basal lineage within Himalayapsitta cluster, P. cyanocephala-finschii were a sister clade to P. roseate based on cytochrome-b gene, RAG-1 (Braun et al. Citation2019). The complete mitochondrial genome of P. roseata has not been determined and characterized until now. Therefore, the aim of this study was to first assemble and characterize the complete mitochondrial genome of P. roseata.
The specimen (Duan-001) was collected from Dehong Wildlife rescue center (24.37 N, 98.48E), which was located southwestern of Yunnan Province in China, and stored at the Herbarium of Southwest Forestry University. The total mitochondrial DNA was extracted from the muscle tissue. The mitogenome was sequenced using Illumina Novaseq sequencing platform (Illumina, San Diego, CA) and assembled with A5-miseq v20150522 (Coil et al. Citation2015) and SPAdesv3.9.0 (Bankevich et al. Citation2012). The complete mitochondrial genome was annotated using MITOS Web server (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). The complete mitochondrial genome of P. roseata was submitted to the NCBI database under the accession number MK986661. Phylogenetic tree of the relationships among 24 species were presented using maximum likelihood (ML) methods available on the CIPRES Science Gateway v3.3 (Miller et al. Citation2010). Bayesian inference was calculated with MrBayes3.1.2 with a general time reversible (GTR) model of DNA substitution and a gamma distribution rate variation across sites (Ronquist and Huelsenbeck Citation2003). Sequences of Psittaciformes (Melopsittacus undulatus and Agapornis roseicollis) obtained frofm GenBank (NC_009134 and NC_011708) were used as outgroups to root trees following Kundu et al. (Citation2012).
The complete mitochondrial genome of P. roseata was found to be a circular double-stranded 16,814 bp in length. A total of 39 mitochondrial genes were identified, including 14 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 non-coding control region (D-loop). All the 14 protein coding genes contained the same start condon ATG, except that cox1 gene started with GTG, nad3_0 gene started with GCC. Furthermore, eleven of the PCGs used complete TAA or incomplete T(AA)/TA(A) stop codon, and other three types were AGG[cox1], AGT[nad3_1], GAA[nad3_0]. The length of 22 tRNA genes ranged from 65 bp (trnF) to 76 bp (trnS2). The 12S rRNA and 16S rRNA genes were located between the trnF and trnL2 genes and separated by the trnV gene, with a length of 976 and 1584 bp, respectively. The D-loop length was 425 bp and laied between the trnE and trnF genes.
Among these genes, nad6 and 8 tRNAs (trnQ, trnA, trnN, trnC, trnY, trnS2, trnP and trnE) were located on the light strand (Lstrand), while all of the remaining genes were located on the heavy strand (H-strand). The overall base composition of P. roseata mitogenome was 31.88% for A, 22.00% for T, 32.88% for C, and 13.23% for G, A + T content is 53.88%, which is higher than G + C content of 46.11%, similar to other Psittaciformes (Liu et al. Citation2019, Citation2019).
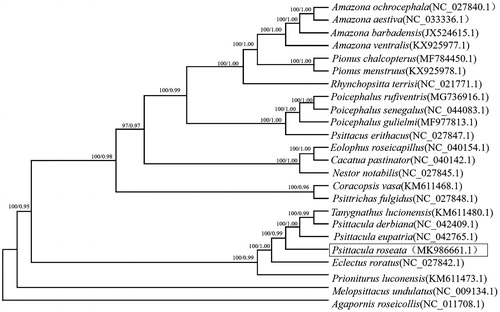
The reconstructed phylogenetic tree showed that P. roseate grouped with P. eupatria, P. derbiana and Tanygnathus lucionensis with strong support (100% bootstrap support value) by the analyses of protein-coding genes (). The genus Psittacula clade was found to be sister to the genus Eclectus, which is congruent with previous studies (Kundu et al. Citation2012; Liu et al. Citation2019). The complete mitochondrial genome of P. roseate reported here will be useful for future population genetic studies of this species and will provide essential genome resources for the ecologically important species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MK986661], or available from the corresponding author.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- BirdLife International. 2019. Species factsheet: Psittacula roseate. [accessed 2019 Oct 5]. http://www.birdlife.org.
- Braun MP, Datzmann T, Arndt T, Reinschmidt M, Schnitker H, Bahr N, Sauer-Gürth H, Wink M. 2019. A molecular phylogeny of the genus Psittacula sensu lato (Aves: Psittaciformes: Psittacidae: Psittacula, Psittinus, Tanygnathus, †Mascarinus) with taxonomic implications. Zootaxa. 4563(3):zootaxa.4563.3.8–562.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589.
- IUCN. 2019. The IUCN Red List of Threatened Species (Version 2019-1). [accessed 2019 Oct 10]. www.incnredlist.org.
- Juniper T, Parr M. 1998. Parrots: a guide to the parrots of the world. Robertsbridge (UK): Pica Press
- Kundu S, Jones CG, Prys-Jones RP, Groombridge JJ. 2012. The evolution of the Indian Ocean parrots (Psittaciformes): extinction, adaptive radiation and eustacy. Mol Phylogenet Evol. 62(1):296–305.
- Liu H, Ding Y, Chen R, Tong Q, Chen P. 2019. Complete mitogenomes of two Psittacula species, P. derbiana and P. Eupatria. Mitochondrial DNA Part B. 4(1):692–693.
- Liu HY, Jin K, Li L. 2019. The complete mitochondrial genome of the Fischer’s Lovebird Agapornis fischeri (Psittaciformes: Psittacidae). Mitochondrial DNA Part B. 4(1):1217–1218.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. New Orleans (LA): Gateway Computing Environments Workshop (GCE), p. 1–8.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574.