Abstract
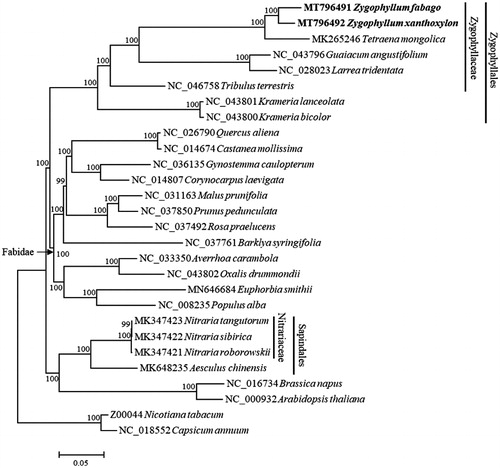
Zygophyllum xanthoxylon and Z. fabago are two important desert plants from Zygophyllaceae, which are both widely distributed in north-western China. Here, we report the complete chloroplast genome sequences of Z. xanthoxylon and Z. fabago, which are 109,577 bp and 108,695 bp in length, respectively. The inverted repeat regions, the large single-copy region and the small single-copy region of Z. xanthoxylon are 5084 bp, 83,735 bp, and 15,674 bp in length, respectively, while those of Z. fabago are 4669 bp, 82,293 bp and 17,064 bp in length, respectively. A total of 98 genes were annotated in the genome of Z. xanthoxylon including 29 tRNA, 4 rRNA and 65 protein-coding genes, and 100 genes were annotated in the genome of Z. fabago including 31 tRNA, 4 rRNA and 65 protein-coding genes. Phylogenetic analysis showed Z. xanthoxylon clustered and Z. fabago formed a monophyletic group sister to Tetraena mongolica.
The genus Zygophyllum are widely distributed in the arid and semi-arid areas of Africa, Europe, Asia and Australia. In the desert of north-western China, the shrub Zygophyllum xanthoxylon and the herb Z. fabago have ever been proposed as members of two distinct genera, Sarcozygium and Zygophyllum, in Zygophyllaceae (Liu Citation1998). However, phylogenetic evidence from chloroplast gene rbcL and trnL-F (Han et al. Citation2016) supported that Sarcozygium was merged into Zygophyllum (Liu and Zhou Citation2008). The Zygophyllaceae species including Zygophyllum and Tetraena are important constituents of local plant community (Wan et al. Citation2006; Ma et al. Citation2019), with key ecological functions on sand fixation and preventing desertification. To better understand the phylogenetic relationship and evolution of the species in Zygophyllaceae, we sequenced the complete chloroplast genome of Z. xanthoxylon and Z. fabago and compared them with the related species.
The DNA of Z. xanthoxylon and Z. fabago was extracted from the fresh leaves which were collected on the north-western Ordos plateau in Inner Mongolia, China. The voucher specimens were deposited in the herbarium of Fudan University (FUS). Fresh leaves dried with silica-gel were used to extract total genomic DNA, which sequenced using the Illumina Hiseq 2500 (Illumina, San Diego, CA, USA), with 150 bp paired-end sequencing. The two complete chloroplast genomes were assembled by SOAPdenovo2 v2.04 (Luo et al. Citation2012) and Bowtie2 v2.3.4.1 (Langmead and Salzberg Citation2012). GapCloser v2.04 (Luo et al. Citation2012) was used to fill the gaps; MUMmer v3.23 (Kurtz et al. Citation2004) was used for linear alignment; and DOGMA was used for annotation (Wyman et al. Citation2004). The two complete chloroplast genome sequences have been submitted to NCBI (https://www.ncbi.nlm.nih.gov, GenBank accession numbers: MT796491 and MT796492). Using RAxML (Stamatakis Citation2014), a maximum likelihood (ML) analysis was conducted for eight species from Zygophyllales including Z. xanthoxylon and Z. fabago, and 20 representative species from nine orders.
The total length of the complete chloroplast genome of Z. xanthoxylon is 109,577 bp and that of Z. fabago is 108,695 bp. In Z. xanthoxylon, the typical quadripartite structure includes an LSC region of 83,735 bp, an SSC region of 15,674 bp and a pair of IRs of 5084 bp. In Z. fabago, the length of corresponding regions are 82,293 bp, 17,064 bp and 4669 bp, respectively. The GC content of two chloroplast genomes are 33.77% and 33.73%. A total of 98 predicted genes including 65 protein-coding genes, 29 tRNA genes and 4 rRNA genes were identified in Z. xanthoxylon and a total of 100 predicted genes including 65 protein-coding genes, 31 tRNA genes and 4 rRNA genes were identified in Z. fabago.
The phylogenetic analysis showed that Z. xanthoxylon and Z. fabago formed a monophyletic group which cluster with T. mongolica (). Besides, our result confirmed that Zygophyllaceae had a closer relationship with other Fabidae taxa than with Nitrariaceae in Sapindales (Dong et al. Citation2019).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank at https://www.ncbi.nlm.nih.gov, reference number MT796491 and MT796492. Illumina raw sequencing data for two Zygophyllum species have been deposited in the NCBI Sequence Read Archive (SRA) under accession SRR12506738 and SRR12506739.
Additional information
Funding
References
- Dong Q, Hu N, Suo Y, Chi X, Wang H. 2019. The complete chloroplast genome sequences of two species from Nitraria. Mitochondrial DNA B. 4(1):1229–1230.
- Han L, Wang SM, Wang JC. 2016. Phylogenetic status and character evolution of Zygophyllum (Zygophyllaceae) species in China and the world. Arid Zone Res. 33:1226–1234.
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Liu Y. 1998. Zygophyllaceae. In: Xu L, Huang C, editors. Flora reipublicae popularis sinicae. Vol. 43(1). Beijing: Scientific Press; p. 116–145.
- Liu Y, Zhou L. 2008. Zygophyllaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 11. St. Louis (MO): Missouri Botanical Garden; p. 41–50.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: and empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18.
- Ma X, Chang J, Li Z, Zhai W, Yu x, Feng Y. 2019. The complete chloroplast genome of Tetraena mongolica (Zygophyllaceae), an endangered shrub endemic to China. Mitochondrial DNA B. 4(1):1030–1031.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wan T, Yan L, Shi XS, Yi WD, Zhang XM. 2006. Comparative analysis of genetic diversity of Zygophyllum L. and its related congener Sarcozygium xanthoxylon Bunge in Inner Mongolia. J Arid Land Resour Environ. 20:199–203.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.