Abstract
Vicia costata Ledeb. is a fine leguminous pasture which has advantages of drought resistance and barren tolerance. In this study, the complete chloroplast (cp) genome of V. costata was assembled and annotated. The total genome size of V. costata was 134,184 bp in length with IR loss. The cp genome encoded a set of 112 genes, containing 74 protein-coding genes, 34 tRNA genes, and 4 rRNA genes. The overall GC content was 34.82%. Phylogenetic analysis showed that V. costata got together with the same genus species V. ramuliflora, V. bungei, V. faba, V. sativa and V. sepium with high support value, and V. costata had a close relationship with V. ramuliflora. The whole cp genome of V. costata will be a useful resource for future studies on phylogeny and conservation in Vicia.
Papilionoideae is the largest and economically the most important subfamily in Fabaceae. Vicia is a medium-sized genus in Papilionoideae and is widely distributed in the temperate zone of the northern hemisphere (Hanelt and Mettin Citation1989). Vicia costata Ledeb. is a typical and precious perennial grass of Vicia. It has the advantages of strong drought resistance, wide adaptability, fast growth speed, high nutritional value, long utilization period, etc., and the first choice of plants for wind prevention and sand fixation, conservation of water and soil and ecological restoration in arid and semi-arid areas. The inverted-repeat-lacking clade (IRLC) including Vicia often exists diversity of changes in structural rearrangements when compared to other angiosperms (Lavin et al. Citation1990; Sabir et al. Citation2014; Li et al. Citation2018; Xin and Yang Citation2020). Owing to its highly conservative structure and low evolutionary rate, the whole chloroplast genomes have become valuable resources for molecular phylogeny and species identification in recent studies. Here, we sequenced the complete chloroplast genome of Vicia costata Ledeb. to enhance our understanding of its genomic information and evolution processes.
Fresh leaves of V. costata were collected from Toketo county test base, Hohhot, Inner Mongolia, China (40°30′25.70″N, 111°24′08.62″E, June 2019). Voucher specimen (no. VICA-P017) was deposited in the laboratory of Inner Mongolia Academy of Agricultural and Animal Husbandry Sciences. Total genome DNA was extracted with the Ezup plant genomic DNA prep kit (Sangon Biotech, Shanghai, China). Total DNA was used to generate libraries with an average insert size of 350 bp. The constructed library was sequenced PE150 by Illumina Hiseq X ten platform. Approximately, 19.6 GB of raw data were generated with 150 bp paired-end read lengths. Then, the raw data were used to assemble the complete cp genome using GetOrganelle software (Jin et al. Citation2019) with Vicia faba (KF042344) as the reference. Genome annotation was performed with the program Geneious R8 (Biomatters Ltd, Auckland, New Zealand) by comparing the sequences with the cp genome of V. faba. The tRNA genes were further confirmed through online tRNAscan-SE web servers (Schattner et al. Citation2005). Where necessary, the positions of start and stop codons and boundaries between introns and exons were manually corrected.
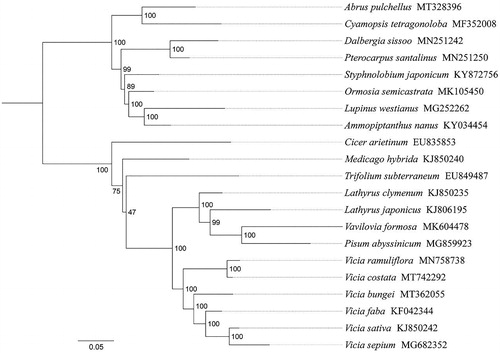
The annotated complete cp genome of V. costata was deposited in GenBank under the accession no. MT742292. The complete chloroplast genome of V. costata was 134,184 bp long with IR loss. The cp genome encoded a set of 112 genes, containing 74 protein-coding genes (PCGs), 34 tRNA genes, and four rRNA genes. The overall GC content was 34.8%. To investigate the phylogenetic position of V. costata, the cp genome sequences of 21 Papilionoideae species were aligned with MAFFT version 7 (Katoh and Standley Citation2013). Then, a maximum likelihood (ML) tree was inferred using raxmlGUI 1.5 (Silvestro and Michalak Citation2012), with the combined rapid bootstrap (1000 replicates) and GTRGAMMA model was used in the ML analysis (). Results showed that the new sequenced species V. costata got together with the same genus (Vicia) species V. ramuliflora, V. bungei, V. faba, V. sativa and V. sepium with high support value (BS = 100), and V. costata had a close relationship with V. ramuliflora. The current study showed that the structure of V. costata cp genome was similar to other Vicia species with inverted repeat loss. These results were largely consistent with previous studies (Li et al. Citation2018; Xin and Yang Citation2020). The data will provide a useful resource for studying the genetic diversity of V. costata, the phylogenetic relationships of the Fabaceae family, and conservation of this valuable species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI (National Center for Biotechnology Information) at https://www.ncbi.nlm.nih.gov/, reference number MT742292.
Additional information
Funding
References
- Hanelt P, Mettin D. 1989. Biosystematics of the genus Vicia L. (Leguminosae). Annu Rev Ecol Syst. 20(1):199–223.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lavin M, Doyle JJ, Palmer JD. 1990. Evolutionary significance of the loss of the chloroplast-DNA inverted repeat in the Leguminosae subfamily Papilionoideae. Evolution. 44(2):390–402.
- Li C, Zhao Y, Huang H, Ding Y, Hu Y, Xu Z. 2018. The complete chloroplast genome of an inverted-repeat-lacking species, Vicia sepium, and its phylogeny. Mitochondr DNA B. 3(1):137–138.
- Sabir JSM, Schwarz EN, Ellison N, Zhang J, Baeshen NA, Mutwakil M, Jansen RK, Ruhlman TA. 2014. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol J. 12(6):743–754.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:686–689.
- Silvestro D, Michalak I. 2012. RaxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337.
- Xin C, Yang Q. 2020. The first complete chloroplast genome sequence of Vicia ramuliflora (Fabaceae). Mitochondr DNA Part B. 5(1):410–411.