Abstract
Saccharina latissima is a brown algal (class Phaeophyceae) belonging to the family Laminariaceae. We reported the de novo assembly and the annotation of the complete chloroplast genome of S. latissima. The circled cpDNA of S. latissima is 130,619 bp in length with a large and a small single-copy region (LSC and SSC), separated by two copies of inverted repeats (IRa and IRb). The genome contains 139 protein-coding genes (PCGs), 3 kinds of ribosomal RNAs (rRNAs), and 29 transfer RNAs (tRNAs) genes that are typical of Saccharina cpDNA. A phylogenetic analysis strongly supported the close phylogenetic affinity of S. latissima and Saccharina japonica. The complete cpDNA of S. latissima will provide valuable molecular data for further analysis of evolutionary and conservation genetic resources.
Saccharina latissima, a species that is distributed in Europe and North America, is of utmost ecological and increasingly economic in coastal areas (Stévant et al. Citation2017). As one of the domain species in oceanic forest, it plays an important role in providing shelter and nursey for numerous animals and purifying the offshore seawater(Christie et al. Citation2009). It belongs to the brown algae (Phaeophyceae) in the order Laminariales, an economic group that has been widely cultivated (Casado-Amezúa et al. Citation2019). Recently, the disappearance of the domain kelp species in European ocean forest (patterns of S. latissima recruitment) and degradation of germplasm in kelp culture caused by the close breeding was reported, indicating much work needs to be done on conservation genetics. Despite the mitochondrial genomes of Saccharina genus were recently analyzed (Wang et al. Citation2015), to our knowledge, the chloroplast genome has never been reported for S. latissima. In fact, so far only one cpDNA was uncovered in Saccharina genus (Wang et al. Citation2013). We here report the assembly of the complete cpDNA of S. latissima to supplement data resources for researches of evolutionary and genetic conservation.
The sample of S. latissima was collected from Germany. Both the specimen and DNA (NO. Saccharina-Ye-C14) are deposited in the herbarium of Yellow Sea Fisheries Research Institute (YSFRI), Qingdao, China. Genome data were obtained by Illumina high throughput technology (see our previous re-sequence project of Saccharina genome) (Ye et al. Citation2015). Illumina paired reads of S. latissima were mapped to the S. japonica chloroplast genome (NC_018523, 130,584 in length) using BWA, which produced the SAM file to the following step for generating the consensus using Genious (http://www.genious.com). No gap was found in the mapping result. The finished sequence was also validated by mapping the raw PE reads back to itself. BLAST (Lobo Citation2008), and Genious were used to perform the genome annotation, following the previous work from Wang et al. (Citation2015).
The circled cpDNA of S. latissima is 1,30,619 bp in length and contains 139 protein-coding sequences (CDS), 29 transfer RNA (tRNA) genes, 3 ribosomal RNA (rRNA) genes (5S rRNA, 16S rRNA, and 23S rRNA, two copy for each). All 139 protein-coding genes (PCGs) have typical initiation codons (ATG). The overall GC content is 30.1%, which is well within the normal range of brown cpDNAs. Nucleotide frequency of the H-strand is as follows: T, 34.40%; A, 34.49%; C, 15.71%; and G, 15.39%. The chloroplast of S. latissima encodes 32,309 amino acids, excluding the stop codons. All the 29 typical tRNAs, ranging from 70 bp to 315 bp, possess a complete clover-leaf secondary structure. The rRNAs of the two 5S rRNAs are 109 bp and 99 bp respectively, the two 16S rRNA are 2944 bp and 2945 bp respectively, and the two copy of 23S rRNA genes are both 1479 bp.
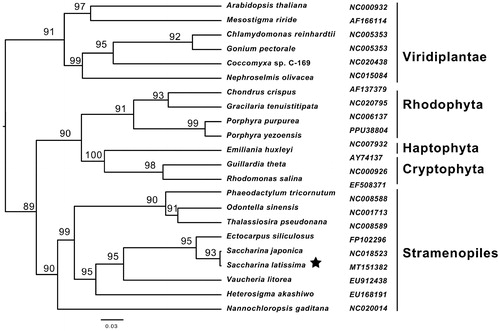
The phylogenetic analyses of the 40 shared proteins were conducted using the maximum likehood method (). The monophyly of three brown seaweeds (Saccharina japonica, S. latissimi and Ectocarpus siliculosus) and those of stramenopiles were clustered with a strong bootstrap value. The MEGA (NJ) analyses yielded the same tree topology. The plastid monophyly of cryptophyta, haptophyta and rhodophyta was significantly clustered with a support value of 90, forming a red-algal clade. Stramenopiles clade and red-algal clade have a closer phylogenetic relationship than the viridiplantae clade, which was widely supported by the secondary endosymbiosis hypothesis (Fan et al. Citation2020).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank with accession MT151382 or China National GeneBank DataBase (CNGBdb) https://db.cngb.org/search/sequence/N_000000748.1/.
Additional information
Funding
References
- Casado-Amezúa P, Araújo R, Bárbara I, Bermejo R, Borja Á, Díez I, Fernández C, Gorostiaga JM, Guinda X, Hernández I, et al. 2019. Distributional shifts of canopy-forming seaweeds from the Atlantic coast of Southern Europe. Biodivers Conserv. 28(5):1151–1172.
- Christie H, Norderhaug KM, Fredriksen SJMEP. 2009. Macrophytes as habitat for fauna. Mar Ecol Prog Ser. 396:221–243.
- Fan X, Qiu H, Han W, Wang Y, Xu D, Zhang X, Bhattacharya D, Ye N. 2020. Phytoplankton pangenome reveals extensive prokaryotic horizontal gene transfer of diverse functions. Sci Adv. 6(18):eaba0111
- Lobo I. 2008. Basic Local Alignment Search Tool (BLAST). J Mol Biol. 215:403–410.
- Stévant P, Rebours C, Chapman A. 2017. Seaweed aquaculture in Norway: recent industrial developments and future perspectives. Aquacult Int. 25(4):1373–1390.
- Wang S, Fan X, Guan Z, Xu D, Zhang XW, Wang DS, Miao Y, Ye NH. 2015. Sequencing of complete mitochondrial genome of Saccharina latissima ye-C14. Mitochondrial DNA, 27(6):1–2.
- Wang X, Shao Z, Fu W, Yao J, Hu Q, Duan D. 2013. Chloroplast genome of one brown seaweed, Saccharina japonica (laminariales, phaeophyta): its structural features and phylogenetic analyses with other photosynthetic plastids. Mar Genomics. 10(Complete):1–9.
- Ye N, Zhang X, Miao M, Fan X, Zheng Y, Xu D, Wang J, Zhou L, Wang D, Gao Y, et al. 2015. Saccharina genomes provide novel insight into kelp biology. Nat Commun. 6:6986–6986.