Abstract
Hippeastrum vittatum (L’Hér.) Herb. is a perennial herb in the Amaryllidaceae, which has been used as a medicinal and ornamental plant. Here, we assembled and characterized the complete chloroplast (cp) genome of H. vittatum by high throughput sequencing. As a result, the length of the complete cp genome is 158,082 bp with a canonical quadripartite structure, consists of a large single-copy region (LSC) of 86,165 bp, a small single-copy region (SSC) of 18,283 bp, and two inverted repeat (IR) regions of 26,817 bp, each. A total of 137 genes were identified, including 87 protein-coding genes, 42 tRNA genes, and 8 rRNA genes. The phylogenomic analysis was performed based on the complete cp genomes of 30 species, which revealed the closest relationship between H. vittatum and H. rutilum in the genus Hippeastrum.
The genus Hippeastrum Herb. contains abundant alkaloids, and is becoming relevant options for neurological disorders and neurodegenerative diseases (Silva et al. Citation2008). It comprises more than 70 species, distributed in tropical and subtropical regions of South America (Poggio et al. Citation2007). There were various hybrids of Hippeastrum plants, and most of the modern commercial hybrids were derived from Hippeastrum vittatum (L’Hér.) Herb. (Robert et al. Citation2006; Phuong et al. Citation2014). Here, we assembled the complete chloroplast genome of H. vittatum by high throughput sequencing, and analyzed the characteristic of the cp genome. Our result will provide more information for the phylogenetic reconstruction of the genus Hippeastrum.
Hippeastrum vittatum was planted in Xinyang Agricultural and Forestry University (E114_13, N32_17), Xinyang, Henan, China. Specimens (no. SZG00057319) were stored at the herbarium of Fairylake Botanical Garden, Shenzhen Chinese Academy of Sciences. Fresh leaves were collected and stored in liquid nitrogen, then stored at −80 °C. Total DNA was isolated using the Plant Genomic DNA Kit (Huayueyang, Beijing, China). DNA integrity and concentration were detected by electrophoresis in 1% (w/v) agarose gel and NanoDrop spectrophotometer 2000. Qualified DNA was used for library construction and sequencing on the DNASEQ T7 platform. A total of 45GB raw data (paired-end 150) was generated, and 117 thousand reads were aligned to the chloroplast genome by the organelle assembler NOVOPlasty Version 3.3 (Dierckxsens et al. Citation2016). Hippeastrum rutilum cp genome sequence (MT133568.1) was chosen as a reference. Finally, genome annotation and visualization were conducted on web server CPGAVAS2 (http://www.herbalgenomics.org/cpgavas2; Shi et al. Citation2019). The complete chloroplast genome sequence of H. vittatum was deposited in GenBank (accession no. MT762362).
The size of H. vittatum cp genome was 158,082 bp with 37.9% GC content. It formed a typical canonical quadripartite structure, including a large single-copy (LSC) region of 86,165 bp, a small single-copy (SSC) region of 18,283 bp, and two inverted repeat (IR) regions of 26,817 bp. A total of 137 genes were annotated, which consisted of 87 protein-coding genes, 42 tRNAs, and 8 rRNAs. Analysis of introns and exons found that there were 17 genes were splitting genes with introns and exons, ycf3 and clpP contain two introns and three exons, others have one intron and two exons, which is the same with some Lycoris species, such as L. radiata (Zhang, Shu, et al. Citation2019) and L. longituba (Zhang, Tong, et al. Citation2019).
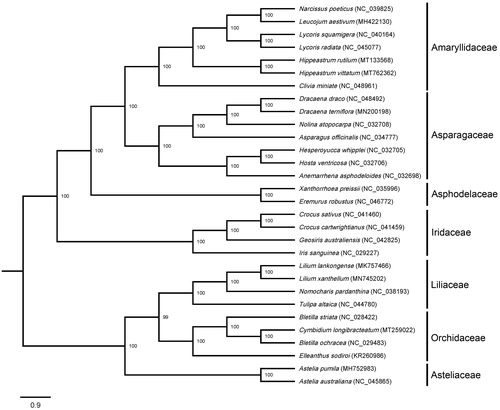
A phylogenetic tree was constructed using 30 species distributed in seven families (). Their cp genome sequences were downloaded from the NCBI GenBank database, then aligned by MAFFT (Rozewicki et al. Citation2019). Maximum-likelihood (ML) phylogeny based on the best-fit model of TVM + F+R4 was conducted using IQ-TREE v. 2.0.3 (Nguyen et al. Citation2015). The best-fit model was chosen by ModelFinder (Kalyaanamoorthy et al. Citation2017) according to the Bayesian information criterion (BIC). The robustness of the topology was estimated using 1000 bootstrap replicates. The result showed the closest relationship between H. vittatum and H. rutilum in the genus Hippeastrum, which was grouped on one branch and near to the genus Lycoris in the Amaryllidaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available at https://www.ncbi.nlm.nih.gov/, accession number (MT762362). The raw sequencing data was uploaded in SRA with the reference number PRJNA659134.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucl Acids Res. 45:e18.
- Kalyaanamoorthy S, Bui MQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Nguyen L, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Phuong Pham TM, Shiro I, Ikuo M. 2014. Genetic variation of Hippeastrum accessions in Vietnam. J Fac Agr Kyushu Univ. 59:235–241.
- Poggio L, Gonzalez G, Naranjo CA. 2007. Chromosome studies in Hippeastrum (Amaryllidaceae): variation in genome size. Bot J Linn Soc. 155(2):171–178.
- Robert B, Jordheim M, Kiremire B, Namukobe J, Andersen ØM. 2006. Anthocyanins from flowers of Hippeastrum cultivars. Sci Hortic-Amsterdam. 109(3):262–266.
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucl Acids Res. 47(W1):W5–W10.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucl Acids Res. 47(W1):W65–W73.
- Silva AFS, De Andrade JP, Machado KRB, Rocha AB, Apel MA, Sobral MEG, Henriques AT, Zuanazzi JAS. 2008. Screening for cytotoxic activity of extracts and isolated alkaloids from bulbs of Hippeastrum vittatum. Phytomedicine. 15(10):882–885.
- Zhang F, Shu X, Wang T, Zhuang W, Wang Z. 2019. The complete chloroplast genome sequence of Lycoris radiata. Mitochondrial DNA B. 4(2):2886–2887.
- Zhang F, Tong H, Yang H, Wang T, Zhuang W, Shu X, Wang Z. 2019. Characterisation of the complete chloroplast genome of Lycoris longituba (Amaryllidaceae). Mitochondrial DNA B. 4(2):3782–3783.