Abstract
Dendrobium wangliangii is a rare orchid species with extremely small populations, endemic to China. In this study, the complete chloroplast (cp) genome sequence and the genome features of D. wangliangii were analyzed. The whole cp genome sequence of D. wangliangii is 160,052 including a large single-copy region (LSC, 87,525 bp), a small single-copy region (SSC, 18,373 bp), and a pair of repeat regions (IRs, 27,077 bp, each). The contents of four bases in cpDNA were A (30.9%), C (18.9%), G (18.3%) and T (31.9%), respectively. The total content of GC is 37.1%. The cp genome contains 129 genes, consisting of 124 unique genes (78 protein-coding genes, 38 tRNAs, and 8 rRNAs). Phylogenetic analysis showed that D. wangliangii nested with other Dendrobium spp. and was closely related to D. ellipsophyllum, D. wattii and D. longicornu.
Dendrobium is one of the largest genera in the orchid family. It consists of approximately 1500 species, mainly distributed in tropical and subtropical regions of the world (Pridgeon et al. Citation2014). Seventy-eight species of this genus in 14 sections were recorded in China (Zhu et al. Citation2009). However, molecular phylogenies of the combination of partial chloroplast marks and ITS-rDNA are not always consistent to morphological sections (Xiang et al. Citation2016). Additional molecular marks such as complete chloroplast genome sequences are needed for phylogenetic and evolutionary analyses.
Dendrobium wangliangii was discovered as a new species in section Dendrobium in 2008 (Hu et al. Citation2008). Dendrobium wangliangii is distributed in specific habitats with narrow distributions, e.g. the Jinshajiang dry-hot valley in Luquan County, Kunming, Yunnan Province (Wang et al. Citation2019), and the populations are extremely small. Given the special features of morphology and the nature of drought tolerance, phylogenetic position of D. wangliangii is unknown. In this study, a complete chloroplast genome (cp genome) sequence of D. wangliangii was characterized and analyzed for its phylogenetic position.
The leave samples of D. wangliangii was collected in Fumin County, Yunnan, China (25°20′19″N, 102° 27′26″E). Total genomic DNA from leaf was extracted using Tiangen DNA kit (TIANGEN, China), and sequenced by the Illumina Hiseq 2500 sequencing platform (Illumina, CA, USA) at Personal Biotechnology Co., Ltd (Shanghai, China). The DNA of D. wangliangii sample was preserved in Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences with collection number JSJSH17122018-1. The sequence data were de novo assembled to a complete and circular cp genome using GetOrganelle (Jin et al. 2020). Gene annotation was accomplished using GeSeq with default sets (Tillich et al. Citation2017), and manually checked in Geneious Primer 2020 (Kearse et al. Citation2012). Sequences of Coding regions (CDS) and rRNAs were extracted, aligned and concatenated in Geneious Primer 2020.
The whole cp genome sequence of D. wangliangii (GenBank accession no. MT798588) is totally 160,052 bp in length, in which the contents of four bases in cpDNA were A (30.9%), C (18.9%), G (18.3%) and T (31.9%), respectively. The total content of GC is 37.1%. The structure of cp genome includes a large single-copy region (LSC, 87,525 bp), a small single-copy region (SSC, 18,373 bp), and a pair of inverted repeat regions (IRs, 27,077 bp, each). Typical code ATG was used as the initiator codon and typical code TAA as the terminator codon in all the 78 protein-coding genes. The gene order and features of the D. wangliangii are consistent with congener species in Dendrobium (Niu, Xue et al. Citation2017; Niu, Zhu, et al. Citation2017).
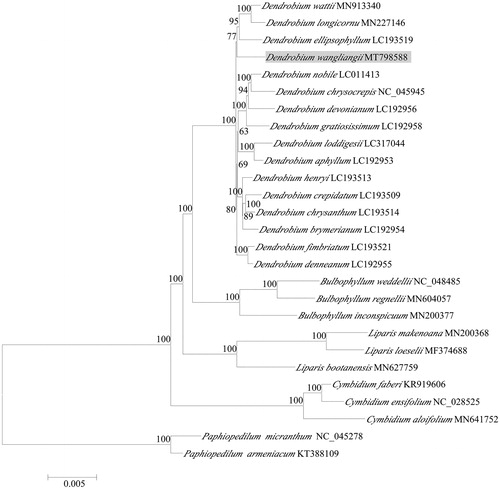
To further investigate its phylogenetic position, the complete chloroplast genomes from 22 species of subfamily Epidendroideae, e.g. 16 Dendrobium species, three Bulbophyllum species and three Cymbidium species, and two species of Paphiopedilum from subfamily Cypripedioideae as outgroup, were selected to reconstruct the phylogenetic tree. The best-fit model GTR + F+R2 using the Akaike information criterion (AIC) was applied to read the best model to construct maximum likelihood (ML) trees using IQ-TREE 1.6 (Lam-Tung et al. Citation2015). Phylogenetic analysis showed that D. wangliangii nested in Dendrobium clade () and was closely related to D. ellipsophyllum, D. wattii and D. longicornu. This newly reported chloroplast genome of D. wangliangii provides important genomic information for Dendrobium species phylogenetic and evolutionary analyses.
Figure 1. Phylogenetic analysis of Dendrobium wangliangii using maximum likelihood of 69 protein-coding genes and four rRNA genes from 16 species from the genus Dendrobium, two species of Paphiopedilum as outgroup. The support values were indicated on the branch node or right side of node; and each tip was labeled by species name and GenBank accession no. The phylogenetic position of the species D. wangliangii was shown in gray box.
Acknowledgments
The authors thank Dr. Wen-Bin Yu for help in software applications and Ms. Bi-Ying Yue for lab assistance during DNA extraction. We are also grateful to Mrs. Xiao-Yun Wang from the Wild Orchid conservation Center of Yunnan Fengchunfang Biotechnology Company for providing sample materials.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the analyses and results of Dendrobium wangliangii in this paper are available in GenBank with accession no. MT798588 (https://www.ncbi.nlm.nih.gov/). Raw sequencing reads was deposited in SRA with BioProject accession: PRJNA659161. (https://www.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA659161).
Additional information
Funding
References
- Hu GW, Long CL, Jin XH. 2008. Dendrobium wangliangii (Orchidaceae), a new species belonging to section Dendrobium from Yunnan, China. Bot J Linn Soc. 157(2):217–221.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2020. Getorganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lam-Tung N, Schmidt HA, von Haeseler A, Bui Quang M. 2015. Iq-tree: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Niu Z, Xue Q, Wang H, Xie X, Zhu S, Liu W, Ding X. 2017. Mutational biases and GC-biased gene conversion affect GC content in the plastomes of Dendrobium genus. Int J Mol Sci. 18:2307.
- Niu Z, Zhu S, Pan J, Li L, Sun J, Ding X. 2017. Comparative analysis of Dendrobium plastomes and utility of plastomic mutational hotspots. Sci Rep. 7:2073.
- Pridgeon AM, Cribb PJ, Chase MW, Rasmussen FN. 2014. Genera Orchidacearum 6. Epidendroideae (part three). Oxford: Oxford University Press.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wang Q, Shao S, Su Y, Hu X, Shen Y, Zhao D. 2019. A novel case of autogamy and cleistogamy in Dendrobium wangliangii: a rare orchid distributed in the dry-hot valley. Ecol Evol. 9(22):12906–12914.
- Xiang XG, Mi XC, Zhou HL, Li JW, Chung SW, Li DZ, Huang WC, Jin WT, Li ZY, Huang LQ, Jin XH. 2016. Biogeographical diversification of mainland Asian Dendrobium (Orchidaceae) and its implications for the historical dynamics of evergreen broad-leaved forests. J Biogeogr. 43(7):1310–1323.
- Zhu GH, Ji ZH, Wood JJ, Wood HP. 2009. Dendrobium. In: Wu CY, Raven PH, Hong DY, editors. Flora of China. Beijing & St. Louis: Science Press & Missouri Botanical Garden Press; p. 367–397.
