Abstract
The complete mitochondrial genome of Ferdinandea cuprea was assembled and annotated applying next generation sequencing, which is the first reported mitogenome reference of species from Rhingiini tribe of Syrphidae family. The mitogenome of F. cuprea has length of 15,907 base pair and comprises of 37 genes (13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes) and one putative control region. Most protein-coding genes are initiated with ATN or CAA(COX1), and terminate with TAA. The results of phylogenetic tree reconstruction statistically suggested the monophyly of Syrphinae, but that of Eristalinae is not supported, and shows tribe Rhingiini is closer to Milesiini.
Syrphidae is one of the most diverse and species-rich clade in Diptera, comprising over 9,600 described species (Thompson Citation2008), play an important ecological role in plant pollinators, natural enemies of plant pests and nutrient recycling(Rotheray and Gilbert Citation2011; Li et al. Citation2017). While Rhingiini is one of the most important tribe of subfamily Eristalinae (Diptera: Syrphidae) (Thompson and Rotheray Citation1998), the systemics and taxonomic classification of this tribe is controversial. Following Peck (1988), the tribe Rhingiini were comprised 11 genera, while Thompson (Citation1972) and Shatalkin (Citation1975) classified the tribe Rhingiini into 10 and 8 genera (Peck Citation1988;Thompson Citation1972; Shatalkin Citation1975; Velterop Citation1986). Ferdinandea cuprea (Scopoli, 1763), a very common hoverfly belonging to the tribe Rhingiini of subfamily Eristalinae (Diptera: Syrphidae) has also an ambiguous status (Scopoli Citation1763; Stuke et al. Citation2004). The significant morphological characteristics are strong black bristles on thorax and scutellum, and dark spots on wings (Thompson and Rotheray Citation1998). The adults of F. cuprea prefer habitat of natural deciduous forests, while larvae mainly live in sap-runs of generally over-mature trees (Ricarte et al. Citation2010). Here, we sequenced and assembled the complete mitogenome of species F. cuprea to contribute more data for better understanding the phylogenetic relationships within the family Syrphidae.
Specimens of F. cuprea were collected from the Changqing National Nature Reserve (107°17′E, 33°19′N) on July 2019 and deposited in the Museum of Zoology and Botany, Shaanxi University of Technology, Hanzhong, China (SUHC), with the accession number of SYY20190077.
Genomic DNA of F. cuprea was extracted using the Qiagen DNeasy kit (Qiagen, Hilden, Germany), and 150 bp paired-end library was constructed for sequencing applying Illumina HiSeq 4000 platform. The raw data of whole genome sequencing have been submitted to the Genome Warehouse in National Genomics Data Center(CNCB-NGDC) (Zhang et al. 2020)with accession number of CRA003174 (https://bigd.big.ac.cn/gsa/). The software MitoZ (Meng et al. Citation2019) was used for mitochondrial genome assembly, annotation and visualization. The putative control region was depended on the boundary of tRNAs. The assembled complete mitogenome of F. cuprea reported in this paper has been deposited in the sequences database of CNCB-NGDC with accession number of GWHAOQX00000000 (https://bigd.big.ac.cn/gwh).
The complete mitogenome of F. cuprea is 15,907 bp in length that includes 37 typical insect mitochondrial genes: 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA), and two ribosomal RNA (rRNA), as well one putative control region (D-loop). No rearrangement was detected in this mitogenome and all genes were arranged in the same order as the putative ancestral arrangement of insects (Cameron, 2014). Twenty-three genes were encoded by the majority strand (J-strand) and the other 14 genes were located by the minority strand (N-strand). With the exception of control region, 8 overlaps (ranging from 2 to 19 bp) and 20 intergenic spacers (1 and 72 bp) were found as intervals between coding genes in this mitogenome. The overall nucleotide composition of the F. cuprea showed a strong AT biased (40.7% of A, 38.9% of T, 8.4% of G, 12.1% of C), a positive AT-skew (0.022) and a negative GC-skew(–0.181) like other Syrphidae mitochondrial genomes (Li et al. Citation2017). Except gene COX1started with CAA, the rest of 12 PCGs use ATN as the initiation codon (ATP8, ND2, ND3, ND5 and ND6 used ATT, COX2, COX3, ND4, ND4L and Cytb used ATG, ATP6 and ND1 used ATA). For all PCGs, TAA are used as termination codon.
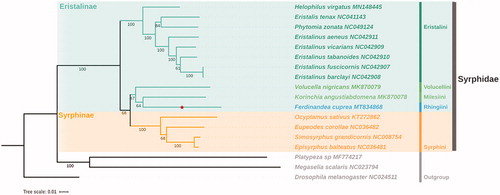
To determine the phylogenetic position of F. cuprea, we reconstructed the phylogenetic tree with other 14 Syrphidae species and three outgroups sequences (available in GenBank database: https://www.ncbi.nlm.nih.gov/). Based on the multiple alignments of 13 PCGs by MAFFT ( Katoh and Standley, Citation2013), the maximum-Likelihood (ML) tree was constructed with IQ-tree (Nguyen et al. Citation2015) with 1000 bootstrap replications (). The tree shows a high support for the monophyly of family Syrphidae which is consistent with the previous published results(Young et al. Citation2016; Li et al. Citation2017) The monophyly of Eristalinae was not supported in present analysis. The subfamily Eristaline, including tribes of Volucellini (V. nigricans), Milesiini (K. angustiabdomena), Rhingiini (F. cuprea) and Eristalini (P. zonata, E. aeneus, E. barclayi, E. fuscicornis, E. tabanoides, E. vicarians, E. tenax, H. virgatus), was not supported as suggested by previous research (Pauli et al. Citation2018), and tribe Rhingiini is closer to Milesiini.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in CNCB-NGDC at https://bigd.big.ac.cn/gwh, reference number GWHAOQX00000000.
Additional information
Funding
References
- Katoh K, Standley D M. 2013. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Li X, Ding S, Li X, Hou P, Tang C, Yang D. 2017. The complete mitochondrial genome analysis of Eristalis tenax (Diptera, Syrphidae). Mitochondrial DNA Part B. 2(2):654–655.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Pauli T, Burt TO, Meusemann K, Bayless K, Donath A, Podsiadlowski L, Mayer C, Kozlov A, Vasilikopoulos A, Liu S, et al. 2018. New data, same story: phylogenomics does not support Syrphoidea (Diptera: Syrphidae, Pipunculidae). Syst Entomol. 43(3):447–459.
- Peck, L. Syrphidae. In: Soos, A., Papp, L. (edutors. Catalogue of Palaearctic Diptera, Vol. 8, Syrphidae–Conopidae. Elsevier, Budapest, Hungary; 1988.
- Ricarte A, Nedeljković Z, Quinto J, Marcos-García MÁ. 2010. The genus Ferdinandea Rondani, 1844 (Diptera, Syrphidae) in the Iberian Peninsula: first records and new breeding sites. J Entomol Res Soc. 12:57–69.
- Rotheray GE, Gilbert F. 2011. The natural history of hoverflies. Cardigan (UK): Forrest Text.
- Scopoli JA.Entomologia carniolica exhibens insecta carnioliae indigena et distribute in ordines, genera, species, varietates, methodo Linnaeana.Trattner, Vindobonae; 1763.
- Shatalkin AI. 1975. A taxonomic analysis of the hover flies (Diptera,Syrphidae). II. Entomol Rev. 54:127–134.
- Stuke JH, Vujić AA, Doczkal D, Muona J, Ståhls G. 2004. Cladistics Rhingiini (Diptera, Syrphidae) based on morphological and molecular characters. Cladistics. 20(2):105–122.
- Thompson FC. 1972. A contribution to a generic revision of the neotropical Milesinae (Diptera: Syrphidae). Arquivos Zool. 23:73–215.
- Thompson FC. 2008. The diptera site – the biosystematic database of world diptera. Version 2.7. [cited 2020 Jul 30]. http://www.sel.barc.usda.gov/diptera/biosys.htm
- Thompson FC, Rotheray G. 1998. Family Syrphidae. In: Papp L, Darva B, editors. Contributions to a manual of palaearctic diptera (with species reference to flies of economic importance). Budapest: Science Herald; Vol. 3, p. 81–139.
- Velterop JHC. 1986. Catalogue of palaearctic diptera. Agric Ecosyst Environ. 16(3–4):299–301.
- Young AD, Lemmon AR, Skevington JH, Mengual X, Ståhls G, Reemer M, Jordaens K, Kelso S, Lemmon EM, Hauser M, et al. 2016. Anchored enrichment dataset for true flies (order Diptera) reveals insights into the phylogeny of flower flies (family Syrphidae). BMC Evol Biol. 16(1):143.
- Zhang Z, Ma L, Abbasi AA, Raza RZ, Cao J, Gao F, Chen R, Gao Y, Zhang C, Yuan L, National Genomics Data Center Members. 2020. Database resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 48:D24–D33.