Abstract
Paphiopedilum emersonii is an endemic terrestrial orchid in China. In this study, the chloroplast genome of P. emersonii was determined from BGISEQ-500 sequencing data. The total chloroplast genome was 162,590 bp in length, consisting of a large single-copy region (LSC, 87,852 bp), a small single-copy region (SSC, 870 bp), and two inverted repeat regions (IRA and IRB, 36,934 bp, each). The complete chloroplast genome contains 131 genes, including 81 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. In addition, the phylogenetic analysis indicates that P. emersonii was sister to Paphiopedilum micranthum. The chloroplast genome will contribute to the research and conservation of P. emersonii.
Paphiopedilum emersonii belongs to the Subfam. Cypripedioideae of Orchidaceae (Chen and Stephan Citation2009). It is native to Guangxi and Guizhou of China (Averyanov et al. Citation2005). Paphiopedilum emersonii is famous for its specialization, large lip, and gorgeous flowers and colors, also known as Slipper Orchid, which has high ornamental value and is one of the most popular Orchidaceae plants in the world (Chen and Stephan Citation2009). Paphiopedilum emersonii has been widely cultivated all over the world, and there is registered in the Royal Horticultural Society (RHS; Zeng et al. Citation2010). However, due to excessive man-made collection, trade, habitat destruction, and other reasons for a long time, the wild resources of P. emersonii has decreased rapidly and even become extinct. All species of Paphiopedilum have been listed in Appendix I of the Convention on International Trade in Endangered species of Wild Fauna and Flora, and international trade is prohibited (Luo et al. Citation2003). Therefore, we report the complete chloroplast genome of P. emersonii, in order to better understand the relationship between P. emersonii and related genera, and contribute to the effective conservation strategy of P. emersonii.
In this study, the complete genomic DNA was extracted from fresh leaves using a modified CTAB method (Doyle and Doyle Citation1987) and sequenced by the BGISEQ-500 platform. The samples were collected from Yachang Orchidaceae National Nature Reserve, Guangxi, China (24°44′N, 106°15′E) and the voucher specimen deposited at the Herbarium of Guangxi Institute of Botany, Guangxi Zhuang Autonomous Region and Chinese Academy of Sciences (specimen code Paphiopedilum_GX).
The clean reads were used to assemble the complete chloroplast genome by the GetOrganelle pipe-line (Jin et al. Citation2018), with the chloroplast genome of Paphiopedilum micranthum (MN_535014) as the reference sequences. The assembled chloroplast genome was annotated using the Geneious R11.15 (Kearse et al. Citation2012). The physical map of the chloroplast genome was generated using the online tool OGDRAW (Lohse et al. Citation2013). Finally, we obtained a complete chloroplast genome of P. emersonii and submitted to GenBank with accession number MT648789.
The total chloroplast genome of P. emersonii was 162,590 bp in length, consisting of a large single-copy region (LSC, 87,852 bp), a small single-copy region (SSC, 870 bp), and two inverted repeat regions (IRA and IRB, 36,934 bp, each). The complete chloroplast genome contained 131 genes, including 81 protein-coding genes, 38 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. The GC content of the chloroplast genome of P. emersonii was 35.6%.
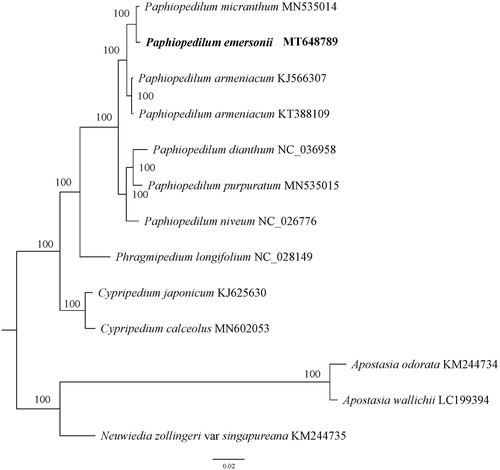
To investigate the phylogenetic position of P. emersonii, a phylogenetic analysis was performed based on 13 complete chloroplast genome sequences of Orchidaceae. All sequences were aligned with the HomBlock pipeline (Bi et al. Citation2018) and subsequently checked manually in Bioedit v5.0.9 (Hall Citation1999). Then, the phylogenetic tree constructed by RAxML (Stamatakis Citation2014) with 1000 ultrafast bootstrap (UFBoot) replicates (Minh et al. Citation2013; Chernomor et al. Citation2016). The results showed that P. emersonii was sister to P. micranthum with 100% bootstrap support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data openly available in a public repository that does not issue DOIs. The data that support the findings of this study are openly available in the National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/), reference number (MT648789).
Additional information
Funding
References
- Averyanov LV, Hiep N, Loc P. 2005. Two relatives of different ecology: a field study of Paphiopedilum emersonii and Paphiopedilum hangianum. Am Orchid Soc Mag. Orchids. 74:208–215.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Chen XQ, Stephan W. 2009. Flora of China. Vol.25. Orchidaceae. Beijing (China): Science Press; St. Louis (MO): Botanical Garden Press. p. 36–37.
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Sym- Posium Series. 41:95–98.
- Jin JJ, Yu WB, Yng JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 256479.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Luo YB, Jia JS, Wang CL. 2003. A general review of the conservation status of Chinese orchids. Biodivers Sci. 11(1):70–77.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zeng SJ, Tian RX, Chen ZL, Wu KL, Duan J. 2010. Research progress on cross breeding of Paphiopedilum. J Tropical Subtropical Bot. 018(004):459–468.