Abstract
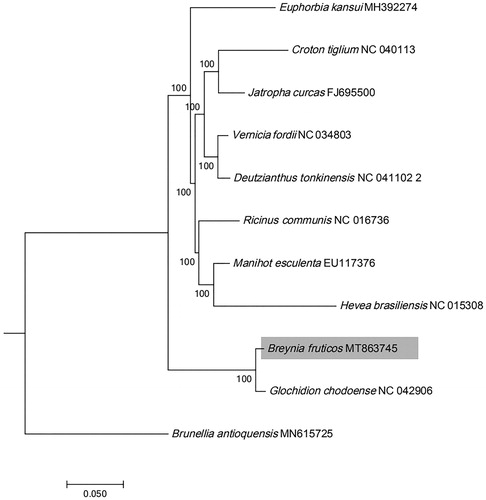
Breynia fruticosa (L.) Müll.Arg. is a well-known folk medicinal plant and found abundantly in South China. The complete chloroplast genome of B. fruticosa reported firstly here was 155,630 bp in length, including a large single-copy region with 85,065 bp (LSC), a small single-copy region with 19,441 bp (SSC) and a pair of inverted repeats with 25, 562 bp (IRa and IRb). The plastome was comprised of 112 distinct genes, with 78 protein coding genes, four ribosomal RNA genes and 30 transfer RNA genes. The overall GC content of B. fruticose chloroplast genome was 36.7%. Phylogenetic analysis revealed that B. fruticosa was closely related to Glochidion fruticosa.
The genus Breynia (Phyllanthaceae) contains about 26 species, and the majority of which are distributed in Southeast Asia (Qiu et al. Citation1996). Four species of Breynia (B. officinalis, B. fruticosa, B. vitis-idaea and B. rostrata) in China are widely used as Chinese traditional medicine (Xie and Xie Citation1996; Sasaki et al. Citation2018). For example, B. fruticosa (L.) Müll.Arg. f., which is widely distributed in South China, is a folk medicine for treatments of gastroenteritis, sore throat, chronic bronchitis and wounds (Meng et al. Citation2010; Liu et al. Citation2011; He et al. Citation2019). Many previous researches have focused on chemical ingredients and medicine activities of Breynia plants (Song Citation1999; Liu et al. Citation2011; Sasaki et al. Citation2018), however there are limited studies on the genetic resource of Breynia species. In this study, the complete plastid genome of B. fruticosa was sequenced and assembled for the first time, and the data obtained here would provide genomic resources of Breynia species for further research.
We collected fresh leaves of of B. fruticosa in Guanghzou, Guangdong province (China; N 23°12′26.02″, E 113°21′5.13″ and deposited the voucher specimen (YAO-YGGD2019040601) in the Herbarium of South China Botanical Garden, Chinese Academy of Sciences (IBSC). Total genomic DNA of B. fruticosa was extracted from its fresh leaves materials using modified CTAB method (Doyle and Doyle Citation1987). Genomic sequencing was performed on the Illumina HisSeq 2500 Sequencing System. Software GetOrganelle (Jin et al. Citation2018) was employed to assemble the reads and PGA (Qu et al. Citation2019) was used to annotate all the plastid genes. The annotated plastid genomic sequence of B. fruticosa is available in the GenBank with the accession number MT863745.
The plastid genome of B. fruticosa was 155,630 bp in length, including a large single-copy region with 85,065 bp (LSC), a small single-copyregion with 19,441 bp (SSC), and a pair of inverted repeats with 25,562 bp (IRa and IRb). The overall GC content of B. fruticose chloroplast genome was 36.7% (LSC: 34.5%, SSC: 30.3%, IRs: 43.0%). The annotated genome predicted 112 distinct genes, including 78 protein coding genes, four ribosomal RNA genes (rrn16, rrn23, rrn4.5, rrn5) and 30 transfer RNA genes. Within the annotated genes, there were 17 genes in the IR regions, with 6 protein-coding genes (rpl2, rpl23, ycf2, ndhB, rps7, rps12), four ribosomal RNA genes (rrn16, rrn23, rrn4.5, rrn5) and seven transfer RNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU).
The maximum likelihood (ML) phylogenetic tree was constructed using RAxML –HPC on the CIPRES cluster. A combined sequence matrix was emoloyed, including all the 78 coding genes of B. fruticosa and Phyllanthaceae and Euphorbiaceae from 9 plastid genomes downloaded from the website of NCBI (www.ncbi.nlm.nih.gov). Sequences were aligned using MAFFT v7.017 plugin in Geneious R11.1.5. The GTR + G model and the default number of rate categories were set. The support values of phylogenetic nodes were conducted on the basis of a rapid bootstrap (BS) analysis with 1000 replicates. The phylogenetic result () showed that Breynia was sister to the genus Glochidion with strong support value (MLBS = 100%). The complete plastid genome of B. fruticosa reported here would provide a useful genetic resource on genetic diversity and conservation of Breynia species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT863745.
Additional information
Funding
References
- Doyle JJ, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- He XL, Lv JJ, Wang X, Zhang Q, Zhang B, Cao K, Liu LL, Xu Y. 2019. The identification and isolation of anti-inflammatory ingredients of ethno medicine Breynia fruticosa. J Ethnopharmacol. 239:111894
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. 256479. doi:10.1101/256479
- Liu YP, Cai XH, Feng T, Li Y, Li XN, Luo XD. 2011. Triterpene and sterol derivatives from the roots of Breynia fruticosa. J Nat Prod. 74:1164–1168.
- Meng DH, Wu J, Zhao WM. 2010. Glycosides from Breynia fruticosa and Breynia rostrata. Phytochemistry. 71(2–3):325–331.
- Qiu HX, Huang SM, Zhang YT. 1996. Flora of China: Euphoribiaceae. In: Qiu HX, Huang SM, Zhang YT editors. Flora of China. Vol. 44, Chapter 1. Beijing, China: Science Press; p. 178–184.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Sasaki A, Yamano Y, Sugimoto S, Otsuka H, Matsunami K, Shinzato T. 2018. Phenolic compounds from the leaves of Breynia officinalis and their tyrosinase and melanogenesis inhibitory activities. J Nat Med. 72(2):381–389.
- Song LR. 1999. Breynia fruticosa, China’s Herbs. Shanghai: Shanghai Scientific & technical publisher; p. 4760–4763.
- Xie ZW, 1996. Brenia fruticose. In Xie ZW, editor. China’s medicinal herbs. Beijing: People’s Health Publishing House; p. 881–882.