Abstract
Astragalus laxmannii is a traditional Chinese medicine. The complete chloroplast genome sequence is 122,844 bp in length, contains 110 complete genes, including 75 protein-coding genes (75 PCGs), 8 ribosomal RNA genes (4 rRNAs), and 30 tRNA genes (30 tRNAs). The overall GC content of cp DNA is 34.1%. Phylogenetic tree shows that A. laxmannii is a sister to A. strictus.
Astragalus species as one of the most important Qi tonifying adaptogenic herbs in traditional Chinese medicine (Yang et al., Citation2010; Zhong et al., Citation2012; Liu et al., Citation2017). It is valued for its ability to strengthen the primary energy of the body which we know as the immune system, as well as the metabolic, respiratory and eliminative functions (Liu et al., Citation2017). This fact is being increasingly substantiated by pharmacological studies showing that it can increase telomerase activity, and has antioxidant, anti-inflammatory, immuneregulatory, anticancer, antitumor, antioxidant, hypolipidemic, antihyperglycemic, hepatoprotective, expectorant, immunomodulatory activity, and diuretic effects (Anon Citation2003; Ma et al. Citation2011; Zhao et al. Citation2011). However, there are very few studies on the A. laxmannii, which greatly limit the development and utilization of A. laxmannii. So far, the chloroplast genome of A. laxmannii has not been reported. In this study, we assembled the complete chloroplast genome of A. laxmannii, hoping to lay a foundation for further research.
Fresh leaves of A. laxmannii were collected from Shapotou (Zhongwei, Ningxia, China; coordinates: 105ast genome of IN\\INdried with silica gel. The voucher specimen was stored in Sichuan University Herbarium with the accstion number of QTPLJQ13383120. Total genomic DNA was extracted with a modified CTAB method (Doyle and Doyle Citation1987) and a 350-bp library was constructed. This library was sequenced on the Illumina NovaSeq 6000 system with 150 bp paired-end reads. We obtained 10 million high quality pair-end reads for A. laxmannii, and after removing the adapters, the remained reads were used to assemble the complete chloroplast genome by NOVOPlasty (Dierckxsens et al. Citation2017). The complete chloroplasts genome sequence of A. nakaianus was used as a reference. Plann v1.1 (Huang and Cronk Citation2015) and Geneious v11.0.3 (Kearse et al. Citation2012) were used to annotate the chloroplasts genome and correct the annotation.
The total plastome length of A. laxmannii (MT786136) is 1,22,844 bp, exhibits a typical quadripartite structural organization, consisting of a large single copy (LSC) region of 66,532bp, two inverted repeat (IR) regions of 20,638 bp and a small single copy (SSC) region of 15,036 bp. The cp genome contains 110 complete genes, including 75 protein-coding genes (75 PCGs), 8 ribosomal RNA genes (4 rRNAs), and 30 tRNA genes (30 tRNAs). The overall GCcontent of cp DNA is 34.1%, the corresponding values of the LSC, SSC, and IR regions are 35.5%, 31.9%, and 43.5%.
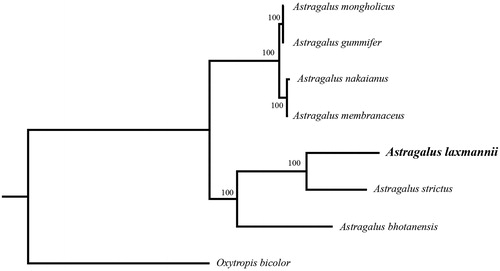
In order to further clarify the phylogenetic position of A. laxmannii, plastome of nine representative Astragalus species were obtained from NCBI to reconstruct the plastome phylogeny, with Oxytropis bicoloras an outgroup. All the sequences were aligned using MAFFT v.7.313 (Katoh and Standley Citation2013) and maximum likelihood phylogenetic analyses were conducted using RAxML v.8.2.11 (Stamatakis Citation2014) under GTRCAT model with 500 bootstrap replicates. The phylogenetic tree shows that the species of Astragaluswere divided into two subclades. A. gummifer, A. mongholicus, A. nakaianus and A. membranaceus clustered together. Remian species clustered in another clade, while A. laxmannii is a sister to A. strictus. ().
Figure 1. Phylogenetic relationships of Astragalus species using whole chloroplast genome. GenBank accession numbers: Astragalus bhotanensis (NC_047381), Astragalus gummifer (NC_047251), Astragalus membranaceus (KX255662), Astragalus mongholicus (NC_029828), Astragalus nakaianus (NC_028171), Astragalus strictus (MT120746), Astragalus laxmannii (MT786136), Oxytropis bicolor (NC_047482).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT786136.
References
- Anon. 2003. Astragalusmembranaceus. Monograph. Altern Med Rev. 8:72–77.
- Bansal D, Bhasin P, Yadav OP, Punia A. 2012. Assessment of genetic diversity in Lepidium sativum (Chandrasur) a medicinal herb used in folklore remedies in India using RAPD. J Genet Eng Biotechnol. 10(1):39–45.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for anno-tating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu P, Zhao H, Luo Y. 2017. Anti-aging implications of Astragalus Membranaceus (Huangqi): a well-known Chinese tonic. Aging Dis. 8(6):868–886.
- Ma J, Qiao Z, Xiang X. 2011. Aqueous extract of Astragalusmongholicus ameliorates high cholesterol diet induced oxidative injury in experimental rat models. J Med Plant Res. 5:855–858.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yang X, Huang S, Chen J, Song N, Wang L, Zhang Z, Deng G, Zheng H, Zhu X-Q, Lu F, et al. 2010. Evaluation of the adjuvant properties of Astragalus membranaceus and Scutellaria baicalensis GEORGI in the immune protection induced by UV-attenuated Toxoplasma gondii in mouse models. Vaccine. 28(3):737–743.
- Zhao LH, Ma ZX, Zhu J, Yu HX, Weng DP. 2011. Characterization of polysaccharide from Astragalu radix as the macrophage stimulator. Cell Immunol. 271(2):329–334.
- Zhong RZ, Yu M, Liu HW, Sun HX, Cao Y, Zhou DW. 2012. Effects of dietary Astragalus polysaccharide and Astragalus membranaceus root supplementation on growth performance, rumen fermentation, immune responses, and antioxidant status of lambs. Anim Feed Sci Technol. 174(1–2):60–67.
