Abstract
Hemerocallis fulva L. is a traditional Chinese medicine. The flowers of H. fulva are used in ethnic medicine to treat various diseases, including certain central nervous system diseases. In this study, we characterized the complete chloroplast genome of H. fulva. It is 156,059 bp in length and encodes 87 protein-coding genes, 38 transfer RNA (tRNA) genes, and 8 ribosomal RNA (rRNA) genes. The phylogenomic analysis showed that the H. fulva and species of Anemarrhena asphodeloides Bunge, Liriope muscari, and Liriope spicata were clustered together. This chloroplast genome sequencing offers genetic background for conservation and phylogenetic studies.
Hemerocallis fulva L.belongs to the Liliaceae family, which is widely used in folk emotional health improvement drugs in East Asia (China, Japan) and North America (Lin et al. Citation2011). Its flower has antioxidation (Lin et al. Citation2011), antibacterial (Sarg et al. Citation1990), antitumor (Cichewicz et al. Citation2004), and sleep improvement effects (Uezu Citation1998). Because of H. fulva flower is rich of hyperin (Guo et al. Citation2013), it is a promising antidepressant drug (Zheng et al. Citation2012).
However, as a result of long-cultivation and interspecific hybrids, there is confusion in the classification of the genus Hemerocallis based on phenotypic characterization. For example, wild Hemerocallis always showed a single flower color, whereas modern hybrid horticultural varieties always showed a more complex color distribution pattern (Cui et al. Citation2019). To provide a scientific classification way to Hemerocallis L., we conducted a chloroplast genome research and a phylogenetic analysis of H. fulva.
Genomic DNA was extracted from fresh leaves of a seedling of H. fulva from Huazhong Medicinal Botanical Garden, Institute of Chinese Medicinal Materials, Hubei Academy of Agricultural Sciences (Hubei, China, N30.180978, E109.756823). Genomic DNA was extracted with plant genomic DNA kit (Tiangen Biotech, China) and sequenced by using the Hiseq 2500 platform (Illumina, San Diego, CA). The chloroplast genome was assembled from the raw sequence data by using NOVOPlasty (v.2.7.2) with the seed sequence of rbcL from Arabidopsis thaliana (Dierckxsens et al. Citation2017). By using Bowtie 2 (v.2.0.1) (Langmead et al. Citation2009) to map all the original reads to the assembly, the correctness of the assembly is verified under the default settings. The annotation of the chloroplast genome was originally performed using CPGAVAS2 (Shi et al. Citation2019) and then edited using Apollo (Misra and Harris Citation2006). The genome sequence and annotations have been deposited in GenBank with accession number MT806177.
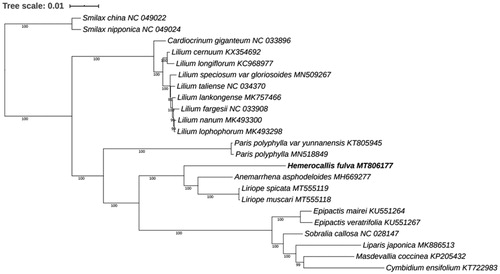
The size of the chloroplast genome of H. fulva is 156,059 bp, including a large single-copy (LSC) region of 84,826 bp and a small single-copy (SSC) region of 18,495 bp separated by a pair identical inverted repeat regions (IRs) of 26,369 bp each. A total of 133 genes were successfully annotated containing 87 protein-coding genes, 38 tRNA genes, and 8 rRNA genes. And the GC content of the three regions is 43, 35, and 32% for IRs, LSC, and SSC, respectively, indicating that IR has the highest GC content. Among them, 11 protein-coding genes had one intron, and 3 protein-coding genes had two introns. 8 tRNA genes were found to contain one intron. To reveal the phylogenetic position of H. fulva with other members of Liliaceae Juss., a phylogenetic analysis was performed based on 21 complete chloroplast genomes from the Liliaceae family. Smilax nipponica (NC_049024) and Smilax china (NC_049022) were set as the outgroups. The MAFFT (7.037 version) (Katoh and Standley Citation2013) was used to extract the coding sequences, and a total of 61 coding sequences (accD, atpA, atpB, atpE, atpF, atpH, atpI, ccsA, cemA, clpP, matK, petA, petB, petD, petG, petL, petN, psaA, psaB, psaC, psaI, psaJ, psbA, psbC, psbD, psbE, psbF, psbH, psbI, psbK, psbL, psbM, psbT, rbcL, rpl2, rpl14, rpl16, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36, rpoA, rpoB, rpoC1, rpoC2, rps2, rps3 rps4, rps7, rps8, rps11, rps12, rps14, rps16, rps18, rps19, ycf2,ycf3, ycf4) were presented in all of the 23 species. Then the MAFFT (7.037 version) was used to concatenate the coding sequences and align the concatenation sequences. RAxML (version 8.2.12) (Stamatakis Citation2014) was used to construct the maximum likelihood (ML) tree; bootstrap probability values were calculated from 1000 replicates. The phylogenetic tree shows that the H. fulva and species of Anemarrhena asphodeloides Bunge, Liriope muscari, and Liriope spicata were clustered together. In this article, we report the complete chloroplast genome of H. fulva, which will provide useful genetic resources for further studying on genetic diversity of this important species and theoretical reference for the classification of Hemerocallis L .
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, the accession number is MT806177, and raw sequencing data used in this study have been deposited in SRA with accession number SRR12506380.
Additional information
Funding
References
- Cichewicz RH, Zhang Y, Seeram NP, Nair MG. 2004. Inhibition of human tumor cell proliferation by novel anthraquinones from daylilies. Life Sci. 74(14):1791–1799.
- Cui H, Zhang Y, Shi X, Gong F, Xiong X, Kang X, Xing G, Li S. 2019. The numerical classification and grading standards of daylily (Hemerocallis) flower color. PLOS One.14(6):e0216460.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:9.
- Guo LQ, Zhang Y, Zhang B. 2013. Chemical constituents and pharmacological research progress of Hemerocallis fulva roots and flowers. J Chin Arch Tradit Chin Med. 31:74–76.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25.
- Lin Y-L, Lu C-K, Huang Y-J, Chen H-J. 2011. Antioxidative caffeoylquinic acids and flavonoids from Hemerocallis fulva Flowers. J Agric Food Chem. 59(16):8789–8795.
- Misra S, Harris N. 2006. Using Apollo to browse and edit genome annotations. Current Protocols in Bioinformatics. Chapter 9:Unit 9.5-Unit 9.5.
- Sarg TM, Salem SA, Farrag NM, Abdel-Aal MM, Ateya AM. 1990. Phytochemical and antimicrobial investigation of Hemerocallis fulva L. grown in Egypt. Int J Crude Drug Res. 28(2):153–156.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Uezu E. 1998. Effects of Hemerocallis on sleep in mice. Psychiatry Clin Neurosci. 52(2):136–137.
- Zheng MZ, Liu CM, Pan FG, Shi DF, Zhang YC. 2012. Antidepressant-like effect of hyperoside isolated from Apocynum venetum leaves: possible cellular mechanisms. Phytomedicine. 19(2):145–149.