Abstract
The mitochondrial genome of the neritid snail Nerita (Heminerita) japonica (Mollusca: Neritimorpha) from Kumamoto, Japan was determined by whole-genome sequencing. This mitogenome is comprised of 13 protein-coding genes, 2 ribosomal RNA (12S and 16S) genes, and 22 transfer RNA genes, with the same gene order as in the other species of the family Neritidae. A likelihood-based phylogenetic reconstruction recovered the subgenus Heminerita (including N. japonica as its type and N. yoldii from China) as monophyletic and sister to a clade with four species of the subgenera Nerita and Theliostyla.
The snails of the genus Nerita (Neritimorpha: Neritidae) are common herbivorous grazers on intertidal rocky shores in tropical, subtropical, and temperate regions worldwide. Approximately, 70 species are identified as the extant members of this genus (Frey and Vermeij Citation2008). A phylogenetic analysis based on partial DNA sequences of a few mitochondrial and nuclear genes has resulted in the recognition of 11 subgenera (Frey Citation2010), whereas relationships among the subgenera remain largely unresolved due to insufficient phylogenetic signal (Frey and Vermeij Citation2008).
The mitochondrial genome (mitogenome) has been assessed for its suitability for the phylogenetic reconstruction of various gastropod taxa (e.g. Uribe et al. Citation2016; Abalde et al. Citation2017; Liu et al. Citation2020). Six species of Nerita belonging to four subgenera have previously been studied in this context (Arquez et al. Citation2014; Feng et al. Citation2019; Xie et al. Citation2019; Castro and Colgan Citation2010). Here, we report the first mitogenome sequence of Nerita (Heminerita) japonica, the type species of Heminerita. This species inhabits upper intertidal rocky shores along the East Asian coast from the mainland Japan to the Korean Peninsula to Zhejiang Province, China (Frey and Vermeij Citation2008; Zhang et al. Citation2018).
A single specimen of N. (H.) japonica was sampled in September 2016 at Shiranui, Uki, Kumamoto, Kyushu Island, Japan (32°38′01″N, 130°38′10″E). Total DNA was extracted from its muscle tissue using a DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) and sequenced on a Miseq System (Illumina, San Diego, CA) at the National Institute for Environmental Studies (Tsukuba, Japan). For de novo assembly in NOVOPlasty 3.7 (Dierckxsens et al. Citation2017), a partial COI fragment of the same specimen was PCR-amplified with the Folmer et al.’s (Folmer et al. Citation1994) primers LCO1490 and HCO2198 and Sanger-sequenced on an ABI 3130xl (Applied Biosystems, Foster City, CA) at Atmosphere and Ocean Research Institute, The University of Tokyo. A total of 3,725,719 paired-end reads from a Miseq run were assembled in NOVOPlasty with the COI sequence as a seed and with the default k-mer value of 39 (Dierckxsens et al. Citation2017). This resulted in a non-circular contig of 15,306 bp. The missing (tRNAGlu and non-coding) region of the mitogenome was amplified with primers designed from both ends of the contig and then sequenced on ABI 3130xl. The amplified sequence had a tandem-repeat region of ca. 250 bp that prevented us to determine a complete, circularized mitogenome, although it added 571 bp to the original contig. The final mitogenome sequence was annotated using the MITOS webserver (Bernt et al. Citation2013) and deposited in the DNA Data Bank of Japan under the accession number LC565707. The specimen was deposited as a voucher (NJ018) in Sesoko Station, University of the Ryukyus.
This 15,877-bp mitogenome sequence contained 13 PCGs, 22 tRNAs, and two rRNAs (12S and 16S). Of the 37 genes identified, seven PCGs and eight tRNAs were encoded on the L-strand, whereas the remaining genes were encoded on the H-strand. The overall base composition was 29.8% for A, 35.4% for T, 21.2% for G, and 13.6% for C with an AT-bias. All PCGs contained ATG as the start codon, and TAA, TAG or T-- as the stop codons. NAD4 and NAD4L genes are overlapped by 7 bp. The 12S (867 bp) and 16S (1,295 bp) rRNA genes were located between tRNALeu and tRNAMet. The lengths of 22 tRNAs ranged from 65 to 74 bp. The gene order was the same as in the previously reported mitogenomes of neritids (Castro and Colgan Citation2010; Arquez et al. Citation2014; Fukumori et al. Citation2016; Uribe et al. Citation2016; Feng et al. Citation2019; Wang et al. Citation2019; Xie et al. Citation2019).
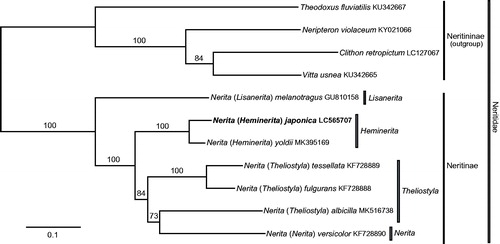
A maximum-likelihood phylogeny of the genus Nerita was inferred from the present and previous mitogenome data, including 13 PCG and two rRNA gene sequences from N. (Heminerita) japonica, N. (H.) yoldii, N. (Theliostyla) albicilla, N. (T.) fulgurans, N. (T.) tessellata, N. (Lisanerita) melanotragus, and N. (Nerita) versicolor. Outgroup comparison was made with four other neritids: Clithon retropictum, Neripteron violaceum, Vitta usnea, and Theodoxus fluviatilis. DNA sequences of each gene were separately aligned using Translator X (for PCGs; Abascal et al. Citation2010) or MAFFT v7 (rDNA; Katoh and Standley Citation2013) with default parameters. Removal of alignment-ambiguous sites (identified by Gblocks v.0.91b (Castresana Citation2000) with all options for a less-stringent selection) resulted in a final dataset of 11 species and 13,394 characters.
The resulting tree () recovered the genus Nerita (and its own subfamily Neritinae; Holthuis Citation1995; Fukumori and Kano Citation2014) as a robust clade with a maximum bootstrap value (100%). The subgenus Heminerita (comprising N. japonica and N. yoldii alone; Frey Citation2010) was monophyletic (100%) and sister to a moderately supported clade of the subgenera Theliostyla and Nerita (84%); the subgenus Theliostyla was rendered paraphyletic to Nerita s.s., albeit with a marginal bootstrap value (73%). The mitogenome seems to provide substantial signal for the phylogenetic reconstruction of the Neritidae at the generic and subgeneric levels.
Figure 1. Maximum likelihood phylogeny of genus Nerita inferred from nucleotide sequences of 13 protein-cording and 2 ribosomal-RNA genes of mitochondrial genome. Tree reconstruction was performed under GTR + G model in RAxML v.7.4.2 (Stamatakis Citation2006) with a bootstrap analysis of 1,000 pseudoreplicates. Mitogenome of Nerita (Heminerita) japonica was newly determined; numbers on branches denote bootstrap values in %.
Acknowledgements
The authors wish to thank Tsuyoshi Takano (Meguro Parasitological Museum) and Genki Kobayashi (Seto Marine Biological Laboratory, Kyoto University) for collecting the specimen of N. (H.) japonica.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in the DNA Data Bank of Japan (DDBJ) at http://getentry.ddbj.nig.ac.jp/top-e.html, reference number LC565707.
Additional information
Funding
References
- Abalde S, Tenorio MJ, Afonso CML, Zardoya R. 2017. Mitogenomic phylogeny of cone snails endemic to Senegal. Mol Phylogenet Evol. 112:79–87.
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38 (Web Server issue):W7–W13.
- Arquez M, Colgan D, Castro LR. 2014. Sequence and comparison of mitochondrial genomes in the genus Nerita (Gastropoda: Neritimorpha: Neritidae) and phylogenetic considerations among gastropods. Mar Genomics. 15:45–54.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Castro LR, Colgan DJ. 2010. The phylogenetic position of Neritimorpha based on the mitochondrial genome of Nerita melanotragus (Mollusca: Gastropoda). Mol Phylogenet Evol. 57(2):918–927.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Feng J, Fu Z, Guo Y, Ye Y, Li J, Guo B, Lü Z. 2019. The complete mitochondrial genome of Nerita albicilla (Neritimorpha: Neritidae). Mitochondrial DNA B Resour. 4(1):1597–1598.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 3:294–299.
- Frey MA. 2010. A revised classification of the gastropod genus Nerita. Veliger. 51:1–7.
- Frey MA, Vermeij GJ. 2008. Molecular phylogenies and historical biogeography of a circumtropical group of gastropods (genus: Nerita): implications for regional diversity patterns in the marine tropics. Mol Phylogenet Evol. 48(3):1067–1086.
- Fukumori H, Itoh H, Kano Y. 2016. The complete mitochondrial genome of the stream snail Clithon retropictus (Neritimorpha: Neritidae). Mitochondrial DNA B Resour. 1(1):820–821.
- Fukumori H, Kano Y. 2014. Evolutionary ecology of settlement size in planktotrophic neritimorph gastropods. Mar Biol. 161(1):213–227.
- Holthuis BV. 1995. Evolution between marine and freshwater habitats: a case study of the gastropod suborder Neritopsina [PhD dissertation]. University of Washington, Seattle, WA.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Liu H, Yang Y, Sun S, Kong L, Li Q. 2020. Mitogenomic phylogeny of the Naticidae (Gastropoda: Littorinimorpha) reveals monophyly of the Polinicinae. Zool Scripta. 49(3):295–306.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Uribe JE, Colgan D, Castro LR, Kano Y, Zardoya R. 2016. Phylogenetic relationships among superfamilies of Neritimorpha (Mollusca: Gastropoda). Mol Phylogenet Evol. 104:21–31.
- Wang P, Zhu P, Wu H, Xu Y, Liao Y, Zhang H. 2019. The complete mitochondrial genome of Neritina violacea. Mitochondrial DNA B Resour. 4(2):2942–2943.
- Xie J, Feng J, Guo Y, Ye Y, Li J, Guo B. 2019. The complete mitochondrial genome and phylogenetic analysis of Nerita yoldii (Gastropoda: Neritidae). Mitochondrial DNA B Resour. 4(1):1099–1100.
- Zhang XJ, Kong LF, Li Q. 2018. DNA barcoding in Neritidae species (Gastropoda, Neritimorpha) along the coast of China. Oceanol Limnol Sin. 49:614–623.
