Abstract
Murraya paniculate, is traditionally used for management of gut, air way and cardiovascular disorders. In this study, we sequenced the complete chloroplast genome of M. paniculata based on next-generation sequencing and used the data to assess genomic resources. The chloroplast genome of M. paniculata is 160,280 bp in length consisting of large and small single-copy regions of length 87,605 and 18,609 bp, separated by two IR regions of 27,033 bp. The overall GC content was 38.61%. De novo assembly and annotation showed the presence of unique genes with 85 protein-coding genes, 29 tRNA genes, and eight rRNA genes. A maximum-likelihood phylogenomic analysis showed that M. paniculata was closely related to M. caloxylon.
Murraya paniculate (Rutaceae), is a folk medicinal plant widely used in Asia as antinociceptive, anti-inflammatory, and antioxidant. (Menezes et al. Citation2017; Sharma et al. Citation2017). However, only a few genomic resources have been explored. Chloroplast genome is valuable sources of genetic markers for phylogenetic analyses, genetic diversity evaluation, and plant molecular identification (Dong et al. Citation2012). Here, we sequenced and analyzed the chloroplast genome of M. paniculata based on the next-generation sequencing method (Dong et al. Citation2013). The main goals of this study were to establish and characterize the organization of the whole chloroplast genome of M. paniculata and to retrieve valuable genomic resources for this species.
We collected fresh healthy leaves from M. paniculata species growing in the Funing, Yunnan province. Voucher specimen was stored in herbarium of Institute of Chinese Materia Medica (CMMI, accession number 532628LY0238), China Academy of Chinese Medical Sciences. Total genomic DNA was extracted and purified following the method of Chen et al. (Citation2019). Paired-end (2 × 150 bp) sequencing was performed by Novogene Bioinformatics Technology Co. Ltd (Beijing, China), using the Illumina Hiseq X-Ten platform. The paired-end reads were qualitatively assessed and assembled with GetOrganelle (Jin et al. Citation2019). The annotation was performed with GeSeq (Tillich et al. Citation2017). The annotated genomic sequence had been submitted to GenBank with the accession number MT747442.
The chloroplast genome of M. paniculata is 160,280 bp in length consisting of large and small single-copy regions of length 87,605 and 18,609 bp, separated by two IR regions of 27,033 bp. GC content was 38.61%. The genome consisted of 122 different genes, including 85 protein-coding genes, 29 distinct tRNA genes, and eight rRNA genes.
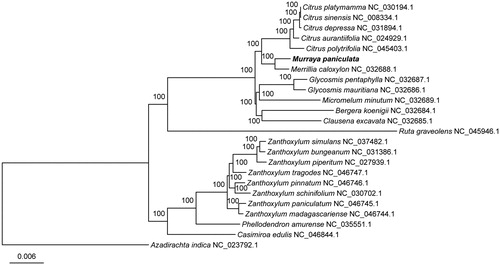
To confirm the phylogenetic location of M. paniculata within the family of Rutaceae, a total of 21 complete cp genomes of Rutaceae were obtained from GenBank, and Azadirachta indica in Meliaceae family was used as out-group. The 23 complete chloroplast sequences were aligned by the MAFFT version 7 software (Katoh and Standley Citation2013). Phylogenetic analysis was conducted based on maximum likelihood (ML) analyses implemented in IQ-TREE 2.0.5 (Minh et al. Citation2020) under the TVM + F + R2 nucleotide substitution model, which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017). Support for the inferred ML tree was inferred by bootstrapping with 1000 replicates. Phylogenetic analysis results strongly supported that M. paniculata was closely related to Merrillia caloxylon (). The chloroplast genome of M. paniculata will provide useful genetic information for further study on genetic diversity and conservation of Rutaceae species.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT747442.
Additional information
Funding
References
- Chen Y, Chen HX, Li HL, Cheng XL, Wang LY. 2019. Complete plastome sequence of Ilex asprella (Hooker and Arnott) Champion ex Bentham (Aquifoliaceae), a Chinese folk herbal medicine. Mitochondrial DNA Part B. 4(2):2341–2342.
- Dong W, Liu J, Yu J, Wang L, Zhou S. 2012. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS One. 7(4):e35071.
- Dong W, Xu C, Cheng T, Lin K, Zhou S. 2013. Sequencing angiosperm plastid genomes made easy: a complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol Evol. 5(5):989–997.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv. 256479.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Menezes CDA, de Oliveira Garcia FA, de Barros Viana GS, Pinheiro PG, Felipe CFB, de Albuquerque TR, Moreira AC, Santos ES, Cavalcante MR, Garcia TR, et al. 2017. Murraya paniculata (L.) (Orange Jasmine): potential nutraceuticals with ameliorative effect in alloxan-induced diabetic rats. Phytother Res. 31(11):1747–1756.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the Genomic Era. Mol Biol Evol. 37(5):1530–1534.
- Sharma P, Batra S, Kumar A, Sharma A. 2017. In vivo antianxiety and antidepressant activity of Murraya paniculata leaf extracts. J Integr Med. 15(4):320–325.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.