Abstract
In this study, the complete mitochondrial genome of Hyphessobrycon herbertaxelrodi is presented, and we also discussed its mitochondrial characteristics. The full length of the mitochondrial genome was 17,417 bp, including 13 protein coding genes (PCGs), 2 ribosomal RNAs (12S and 16S), 22 transfer RNA genes, 1 non-coding control region (D-loop), and 1 origin of replication on the light-strand. The total nucleotide composition of mitochondrial DNA was 29.76%A, 29.88%T, 25.35%C, 15.01%G, and AT was 59.64%. The phylogenetic tree suggested that H. herbertaxelrodi shared the most recent common ancestor with Astyanax giton, Grundulus bogotensis, Astyanax paranae, and Oligosarcus argenteus.
Hyphessobrycon is one of the largest genera in the family Characidae comprising about 130 species distributed from Southern Mexico to the Río de la Plata in Argentina (Carvalho and Francisco Citation2013; Ingenito et al. Citation2013). The black neon tetra (H. herbertaxelrodi) is a well-known freshwater species (Gimeno et al. Citation2016). It is an ornamental fish, and acidic soft water will make the fish show its color. Hyphessobrycon herbertaxelrodi inhabits streams and lakes, likes to swim in groups, has a mild temperament, is omnivorous, and feeds on worms, crustaceans and plants. In this study, we described the complete mitochondrial genome of H. herbertaxelrodi and explored the phylogenetic relationship within Characiformes, to gain its molecular information and thus contribute to facilitate future studies on population genetic structure and phylogenetic relationships.
The sample of H. herbertaxelrodi used in this research was from Zhejiang Oceanography Laboratory (No. 20190825hld35). Total genomic DNA was extracted from a single specimen (SHOUBC72_1) using the improved method with multi-well plates (Pall Corporation) (Yue and Orban Citation2005). Subsequently, based on the existing complete mitochondrial gene of Salminus brasiliensis (KM245047.1) (Brandão-Dias et al. Citation2016), 18 pairs of primers were designed, the samples were amplified by PCR, and then sequenced using Sanger sequencing technology. The complete mitochondrial genome was annotated using Sequin version 15.10 (http://www.ncbi.nlm.nih.gov/Sequin) and tRNAscan-SE version 2.0 (http://trna.ucsc.edu/tRNAscan-SE/) (Patricia and Todd Citation2019). The whole mitochondrial genome of H. herbertaxelrodi was a closed circular molecule composed of 17,147 bp (GenBank accession no. MT769327.1), which was very similar to other typical vertebrate mitochondria (Boore Citation1999; Zhu et al. Citation2018). The complete mitochondrial genome contains 13 protein-coding genes (PCGs), two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNAs (tRNA) genes, and a putative control region (D-Loop) and one origin of replication on the light-strand (OL). The overall base composition is 29.76%A, 29.88%T, 25.35%C, 15.01%G, respectively, with a slight AT bias (59.64%). Most genes of H. herbertaxelrodi mitochondria are encoded on the H-strand, and only ND6 and eight tRNA (Gln, Ala, Asn, Cys, Tyr, Ser, Glu, and Pro) genes are encoded on the L-strand. In the 13PCGs gene, except that the COI starts with GTG, the others use ATG as the start codon, which is very common in vertebrate mtDNA (Prabhu et al. Citation2020). Most of the termination codon TAA or T-, only the stop codon COI for AGG. The lengths of 12S rRNA located between tRNAPhe and tRNAVal and 16S rRNA located between tRNAVal and tRNALeu were 949 bp and 1500 bp, respectively. All 22 tRNAs distributed on the H and L-strands were between 66 and 74 bp in length. 14 tRNA genes were encoded on the H and eight on the L-strands. Most of tRNAs could form a common cloverleaf secondary structure, except tRNASer(AGC) gene without DHU stem (Steinberg et al. Citation1993; Patricia and Todd Citation2019).
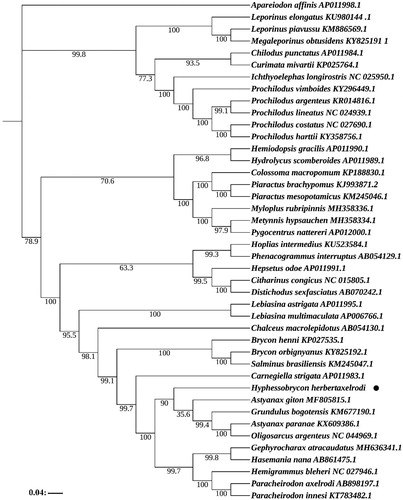
The phylogenetic tree based on the maximum-likelihood (ML) method was constructed to provide relationship within Characiformes. According to the Akaike Information Criteria (AIC), the most suitable nucleotide sequence model was selected through MrModeltest 2.3, and finally the most suitable model was GTR + I+G. The ML phylogenetic tree based on 13 PCGs of 42 species using the software PhyML 3.0 (Guindon et al. Citation2010). The phylogenetic tree suggests that H. herbertaxelrodi shares the most recent common ancestor with Astyanax giton, Grundulus bogotensis, Astyanax paranae and Oligosarcus argenteus ().
Figure 1. Maximum-likelihood (ML) phylogenetic tree based on 13 PCGs was used to study 42 species. The bootstrap values are based on 1000 resamplings. The number at each node was the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labeled with a black dot.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in “NCBI” at https://www.ncbi.nlm.nih.gov/, reference number MT769327.1.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Brandão-Dias PF, Carmo AO, Martins AP, Pimenta RJ, Alves CB, Kalapothakis E. 2016. Complete mitochondrial genome of Salminus brasiliensis (Characiformes, Characidae). Mitochondr DNA A DNA Mapp Seq Anal. 27(3):1577–1578.
- Carvalho FR, Francisco L. 2013. Hyphessobrycon uaiso: new characid fish from the Rio Grande, upper Rio Paraná basin, Minas Gerais State (Ostariophysi: Characidae), with a brief comment about some types of Hyphessobrycon. Neotrop Ichthyol. 11(3):525–536.
- Gimeno E, Quera V, Beltran FS, Dolado R. 2016. Differences in shoaling behavior in two species of freshwater fish (Danio Rerio and Hyphessobrycon Herbertaxelrodi). J Comp Psychol. 130(4):358–368.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321.
- Ingenito LFS, Lima FCT, Buckup PA. 2013. A new species of Hyphessobrycon Durbin (Characiformes: Characidae) from the Rio Juruena basin, Central Brazil, with notes on H. loweae Costa & Géry. Neotrop Ichthyol. 11(1):33–44.
- Patricia PC, Todd ML. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Prabhu VR, Singha HS, Kumar RG, Gopalakrishnan A, Nagarajan M. 2020. Characterization of the complete mitochondrial genome of Barilius malabaricus and its phylogenetic implications. Genomics. 112(3):2154–2163.
- Steinberg S, Misch A, Sprinzl M. 1993. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 21(13):3011–3015.
- Yue GH, Orban L. 2005. A simple and affordable method for high-throughput DNA extraction from animal tissues for polymerase chain reaction. Electrophoresis. 26(16):3081–3083.
- Zhu K, Lü Z, Liu L, Gong L, Jiang L, Liu B. 2018. The complete mitochondrial genome of Hyporhamphus dussumieri (Beloniformes; hemiramphidae) and phylogenetic studies of Beloniformes. Mitochondr DNA B. 3(2):1233–1234.
