Abstract
Amana baohuaensis is a new species that was just named in 2019. Here, we obtained the complete chloroplast (cp) genome of A. baohuaensis using the Illumina paired-end sequencing technology. The cp genome has a typical quadripartite structure with 150,757 bp in length, containing a large single-copy (LSC) region of 81,757 bp, a small single-copy (SSC) region of 16,962 bp, and two inverted repeat (IR) regions of 26,019 bp. The total GC content is 36.73%, of which, the GC content of LSC, SSC and IR regions are 34.63%, 30.11% and 42.20%, respectively. The cp genome of A. baohuaensis contains 111 unique genes, including 78 protein-coding genes, 29 tRNA genes, and four rRNA genes. The Maximum Parsimony (MP) phylogenetic analysis suggested that A. baohuaensis had the closest relationship with A. wanzhensis, and all Amana species grouped together with high bootstrap support.
The genus of Amana is endemic to East Asia, which have six species in China (Tan et al. Citation2007; Han et al. Citation2014; Wang et al. Citation2019). This genus was previously placed in the genus Tulipa. For those genus plants were morphologically distinct from other tulips, they were then separated into a separate genus Amana (Wu et al. Citation2003; Tan et al. Citation2005; Li et al. Citation2017). The morphology of Amana genus plants is similar, and there are some difficulties in classification. With the development of gene sequencing technology, more and more researchers began to focus on the study of chloroplast (cp) genome. The cp genome is generally more conservative (Meng et al. Citation2018), which is of great significance for species identification and phylogeny (Xu et al. Citation2017; Gu et al. Citation2018). Amana baohuaensis B.X. Han, Long Wang & G.Y. Lu is a new species that was just named in 2019 and only occurs in East China, and the wild resource is few (Wang et al. Citation2019). In this study, the complete cp genome of A. baohuaensis was sequenced and analyzed, which provided a reference for systematic evolution and identification of this species.
The fresh leaves of A. baohuaensis were collected in Mt. Baohua, Jurong city, Jiangsu Province, China (32°07′31″ N, 119°05′24″ E). The voucher specimen was deposited in the Center of Herbarium, China Pharmaceutical University, Nanjing, China, under accession number WL194104. Whole genomic DNA was extracted from fresh leaves using Rapid Plant Genomic DNA Isolation Kit, Sangon Biotech (Shanghai) Co., Ltd. Then, the quality of DNA was checked using BioPhotometer Plus (Eppendorf, Germany). The good quality of DNA was sent to GENEWIZ (Suzhou, China) for sequencing. DNA sequencing was conducted on an Illumina Xten platform. Raw sequencing data were deposited in NCBI Sequence Read Archive (SRA) with the accession number PRJNA661717. Clean reads of the matched reference genes were extracted using the cp genome sequence of A. edulis (GenBank No. NC_034707.1) as a reference. These clean reads were assembled using NOVOPlasty v2.7.2 (Dierckxsens et al. Citation2016), and the assembled genome was annotated and analyzed using the online tool GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html), and manually adjusted. Finally, the cp genome sequence was deposited in GenBank with the accession number MT898423.
The complete cp genome of A. baohuaensis is 150,757 bp in length, which had the cyclic tetrad structure, containing a large single-copy (LSC) region of 81,757 bp, a small single-copy (SSC) region of 16,962 bp, and two inverted repeat (IR) regions of 26,019 bp. The total GC content is 36.73%, of which, the GC content of LSC, SSC and IR regions are 34.63%, 30.11% and 42.20%, respectively. A total of 111 genes were annotated in the cp genome of A. baohuaensis, including 78 protein-coding genes, 29 tRNA genes, and four rRNA genes.
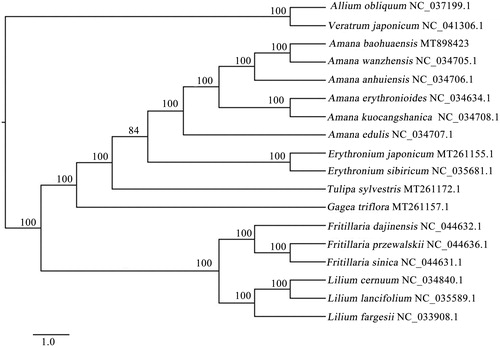
For phylogenetic analysis, the complete cp genomes sequences of 18 species from Liliaceae were used to construct the phylogenetic tree, and Allium obliquum and Veratrum japonicum as outgroups. The sequences were aligned by MAFFT (Katoh and Standley Citation2013), and the sequences alignment result was examined and manually adjusted by BioEdit v. 7.0.9.0 (Hall Citation1999). Maximum Parsimony (MP) analysis was implemented using PAUP* v. 4.0 beta 10 (Swofford Citation2002) with 1000 bootstrap replicates to assess the reliability of the phylogenetic tree. The results showed that A. baohuaensis had the closest relationship with A. wanzhensis, and all Amana species grouped together with high bootstrap support (). This study could provide valuable insight into systematic evolution, conservation and identification for species of Amana.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT898423.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):1–9.
- Gu CH, Tembrock LR, Zheng SY, Wu ZQ. 2018. The complete chloroplast genome of Catha edulis: a comparative analysis of genome features with related species. IJMS. 19 (2):525.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Han BX, Zhang K, Huang LQ. 2014. Amana wanzhensis (Liliaceae), a new species from Anhui, China. Phytotaxa. 177(2):118–124.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Li P, Lu RS, Xu WQ, Ohi-Toma T, Cai MQ, Qiu YX, Cameron KM, Fu CX. 2017. Comparative genomics and phylogenomics of East Asian Tulips (Amana, Liliaceae). Front Plant Sci. 8:451.
- Meng J, Li XP, Li HT, Yang JB, Wang H, He J. 2018. Comparative analysis of the complete chloroplast genomes of four aconitum medicinal species. Molecules. 23(5):1015–1017.
- Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4.0b 10. Sunderland: Sinauer.
- Tan DY, Li XR, Hong DY. 2007. Amana kuocangshanica (Liliaceae), a new species from south-east China. Bot J Linn Soc. 154(3):435–442.
- Tan DY, Zhang Z, Li XR, Hong DY. 2005. Restoration of the genus Amana Honda (Liliaceae) based on a cladistic analysis of morphological characters. Acta Phytotaxon Sin. 43(3):262–270.
- Wang LONG, Xing QIAN, Lu G-YU, Lu XU, Zhao QUN, Song X-W, Han B-X. 2019. Amana baohuaensis (Liliaceae), a new species from East China. Phytotaxa. 427(1):43–50.
- Wu ZY, Lu AM, Tang YC, Chen ZD, Li DZ. 2003. The families and genera of angiosperms in China a comprehensive analysis. Beijing: Science Press; p. 255.
- Xu C, Dong W, Li W, Lu Y, Xie X, Jin X, Shi J, He K, Suo Z. 2017. Comparative analysis of six lagerstroemia complete chloroplast genomes. Front Plant Sci. 8:15.