Abstract
The taxonomic placement of the moth-butterfly, Macrosoma conifera (Warren 1897) (Lepidoptera: Hedylidae), has been controversial. The 15,344 bp complete M. conifera circular mitogenome, assembled by genome skimming, consists of 81.7% AT nucleotides, 22 tRNAs, 13 protein-coding genes, 2 rRNAs and a control region in the typical butterfly gene order. Macrosoma conifera COX1 features an atypical CGA start codon while ATP6, COX1, COX2, and ND5 exhibit incomplete stop codons completed by the post-transcriptional addition of 3′ A residues. Phylogenetic reconstruction places M. conifera as sister to the skippers (Hesperiidae), which is consistent with several recent phylogenetic analyses.
Macrosoma conifera (Warren 1897) is a moth-butterfly species (Lepidoptera: Hedylidae) found in Central and South America (Kawahara et al. Citation2018). It has been most intensively studied in Costa Rica, where its primary larval host plant was identified as Conostegia xalapensis (Melastomataceae), but it has also been observed to feed on Miconia argentea, M. impetiolaris, M. smaragdina (Melastomataceae), Apeiba membranacea and Luehea speciosa (Malvaceae) (Quesada Citation2019). Macrosoma conifera is known for its moth-like features such as nocturnal activity, clubless antennae and dark-adapted visual systems, which distinguish it from the predominantly diurnal butterfly families (Yack et al. Citation2007; Kawahara et al. Citation2018).
There has been considerable controversy regarding the relationship of the moth-butterflies to true butterflies and skippers (Scoble and Aiello Citation1990). At various times, Macrosoma has been associated with moth families Pyralidae and Geometridae; with butterfly families Papilionidae, Pieridae, and Nymphalidae; with skipper family Hesperiidae; and has been proposed as the sister taxon to all skippers and butterflies. These taxonomic placements have been based on either morphological characters (Kendall Citation1976; Scoble Citation1986; Weintraub and Miller Citation1987; Ackery et al. Citation1999; Kristensen and Skalski Citation1999) or phylogenetic analysis of limited numbers of molecular characters (Weller and Pashley Citation1995). More recent molecular phylogenetic analyses have supported a sister relationship between Macrosoma (Hedylidae) and skippers (Hesperiidae) (Wahlberg et al. Citation2005; Regier et al. Citation2009; Mutanen et al. Citation2010; Heikkilä et al. Citation2012; Kawahara et al. Citation2018). Here we report the complete mitochondrial genome sequence of M. conifera from specimen Mcon2013.1, collected in Braulio Carillo National Park, Heredia Province, Costa Rica (GPS 10.1519 N, 84.0945 W) on 2 May 2013 that has been pinned, spread, and deposited in the Wallis Roughley Museum of Entomology, University of Manitoba (voucher WRME0507732).
DNA was prepared (McCullagh and Marcus Citation2015) and sequenced by Illumina NovaSeq6000 (San Diego, California) (Marcus Citation2018). The sequencing library was prepared using NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, Massachusetts). The mitogenome of M. conifera (Genbank MT852025) was assembled by Geneious 10.0.9 from 23,184,240 paired 150 bp reads (Genbank SRA PRJNA662440) using a Mallika jacksoni reference mitogenome (Lepidoptera: Nymphalidae, MT704828 (Alexiuk et al. Citation2020)). Annotation was in reference to M. jacksoni (Lepidoptera: Nymphalidae, MT704828), Kallimoides rumia (Lepidoptera: Nymphalidae, MT704827 (Payment et al. Citation2020)), and Junonia stygia (Lepidoptera: Nymphalidae, MN623383 (Living Prairie Mitogenomics Consortium Citation2020)) mitogenomes. The M. conifera nuclear rRNA repeat (Genbank MT878224) was assembled to a Coeliades ramanatek reference sequence (Genbank MT859413, SRA SRX6097006 (Zhang et al. Citation2019)) and annotated using C. ramanatek, Lerema accius (Genbank MT859412, SRA SRX1085012 (Cong et al. Citation2015)) and Pyrgus malvae (Genbank MT859414, SRA SRX4091999, (Li et al. Citation2019)) (Lepidoptera: Hesperiidae) rRNA repeat sequences.
The M. conifera circular 15,344 bp mitogenome assembly was composed of 3,530 paired reads with nucleotide composition: 40.5% A, 10.9% C, 7.5% G, and 41.2% T. The M. conifera mitogenome gene order and composition is identical to the typical lepidopteran gene arrangement, shared by all families suspected of being closely related to Hedylidae (Park et al. Citation2016). Ten M. conifera mitochondrial protein-coding genes begin with typical ATG or ATT start codons, with the remaining genes beginning with ATA (ND2), ATC (ND6), or CGA (COX1) start codons (Liao et al. Citation2010). The mitogenome contains three protein-coding genes (COX1, COX2, ND5) with single-nucleotide (T) stop codons, and one protein-coding gene (ATP6) with a two-nucleotide (TA) stop codon completed by post-transcriptional addition of 3′ A residues. The locations and structures of tRNAs were determined using ARWEN v.1.2 (Laslett and Canback Citation2008). tRNAs have typical cloverleaf secondary structures except for trnS (AGN) where the dihydrouridine arm is replaced by a loop, while the mitochondrial rRNAs and control region are typical for Lepidoptera (McCullagh and Marcus Citation2015).
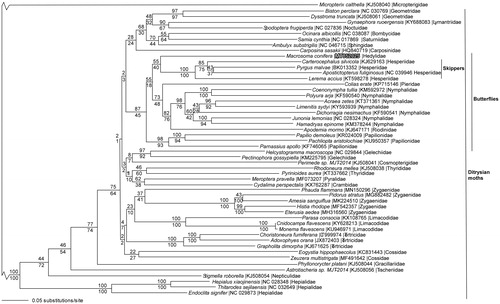
We reconstructed a phylogeny using complete mitogenomes from M. conifera, 50 additional ditrysian Lepidoptera mitogenomes, and a partial mitogenome sequence from basal lepidopteran Micropterix calthella (Lepidoptera: Micropterigidae) as an outgroup (Timmermans et al. Citation2014; Cong and Grishin Citation2016; Lalonde and Marcus Citation2017). Mitogenome sequences were aligned in CLUSTAL Omega (Sievers et al. Citation2011), then analyzed by both parsimony and maximum likelihood (model selected by jModeltest 2.1.7 (Darriba et al. Citation2012), followed by a likelihood ratio test (Huelsenbeck and Rannala Citation1997)) in PAUP* 4.0b8/4.0d78 (Swofford Citation2002) (). Phylogenetic analysis places the swallowtail butterflies (family Papilionidae) as the basal lineage within the butterfly clade (superfamily Papilionoidea), followed by M. conifera and the skippers (family Hesperiidae) as sister taxa. This contradicts several early taxonomic hypotheses, but is consistent with several more recent molecular phylogenetic analyses (Wahlberg et al. Citation2005; Regier et al. Citation2009; Mutanen et al. Citation2010; Heikkilä et al. Citation2012; Kawahara et al. Citation2018). We conclude that the Macrosoma species in family Hedylidae fall within the butterfly clade. Since butterflies are a specialized clade of mostly diurnal moths (Kristensen Citation1999), it is also correct to refer to Macrosoma as moths.
Figure 1. Maximum likelihood phylogeny (GTR + I + G model, I = 0.1590, G = 0.3930, likelihood score 316053.05651) of Marcosoma conifera, 50 additional ditrysian Lepidoptera mitogenomes (including the Pyrgus malvae (Hesperiidae) mitogenome (Genbank BK013352, SRA SRR7174492 (Li et al. Citation2019)), and Micropterix calthella (Micropterigidae)(Timmermans et al. Citation2014) as an outgroup based on 1 million random addition heuristic search replicates (with tree bisection and reconnection). One million maximum parsimony heuristic search replicates produced an identical tree topology (parsimony score 76038 steps). Numbers above each node are maximum likelihood bootstrap values and numbers below each node are maximum parsimony bootstrap values (each from 1 million random fast addition search replicates). Note that the very long branches leading to Micropterix calthella and the basal ditrysian moths are not drawn to scale to facilitate visualing the branching patterns within the ditrysian moths.
Acknowledgments
We thank Genome Quebec for assistance with library preparation and sequencing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference numbers PRJNA662440, MT852025, MT859412-MT859414, MT878224, and BK013352.
Additional information
Funding
References
- Ackery PR, de Jong R, Vane-Wright RI. 1999. The butterflies: Hedyloidea, Hesperioidea and Papilionoidea. In: Kristensen, NP, editor. Lepidoptera: moths and butterflies 1 evolution, systematics and biogeography handbook of zoology. Vol. 4. Berlin: de Gruyter; p. 263–300.
- Alexiuk MR, Marcus JM, Lalonde MML. 2020. The complete mitochondrial genome of the Jackson’s Leaf butterfly Mallika jacksoni (Insecta: Lepidoptera: Nymphalidae). Mitochondr DNA B Resour. 5(3):3316–3318.
- Cong G, Grishin NV. 2016. The complete mitochondrial genome of Lerema accius and its phylogenetic implications. PeerJ. 4:e1546.
- Cong Q, Borek D, Otwinowski Z, Grishin NV. 2015. Skipper genome sheds light on unique phenotypic traits and phylogeny. BMC Genomics. 16:639.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772.
- Heikkilä M, Kaila L, Mutanen M, Peña C, Wahlberg N. 2012. Cretaceous origin and repeated tertiary diversification of the redefined butterflies. Proc Biol Sci. 279(1731):1093–1099.
- Huelsenbeck JP, Rannala B. 1997. Phylogenetic methods come of age: testing hypotheses in an evolutionary context. Science. 276(5310):227–232.
- Kawahara AY, Breinholt JW, Espeland M, Storer C, Plotkin D, Dexter KM, Toussaint EFA, St Laurent RA, Brehm G, Vargas S, et al. 2018. Phylogenetics of moth-like butterflies (Papilionoidea: Hedylidae) based on a new 13-locus target capture probe set. Mol Phylogenet Evol. 127:600–605.
- Kendall RO. 1976. Larval foodplants and life history notes for eight moths from Texas and Mexico. J Lepid Soc. 30:264–271.
- Kristensen NP. 1999. Lepidoptera, moths and butterflies: evolution, systematics and biogeography. Vol 1. New York, USA: Walter de Gruyter.
- Kristensen NP, Skalski AW. 1999. Phylogeny and palaeontology. In: Kristensen, NP, editor. Lepidoptera, moths and butterflies: evolution, systematics and biogeography. Vol. 1. New York, USA: Walter de Gruyter; p. 7–25.
- Lalonde MML, Marcus JM. 2017. The complete mitochondrial genome of the long-horned caddisfly Triaenodes tardus (Insecta: Trichoptera: Leptoceridae). Mitochondr DNA B Resour. 2(2):765–767.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Li W, Cong Q, Shen J, Zhang J, Hallwachs W, Janzen DH, Grishin NV. 2019. Genomes of skipper butterflies reveal extensive convergence of wing patterns. Proc Nat Acad Sci USA. 201821304.
- Liao F, Wang L, Wu S, Li Y-P, Zhao L, Huang G-M, Niu C-J, Liu Y-Q, Li M-G. 2010. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci. 6(2):172–186.
- Living Prairie Mitogenomics Consortium. 2020. The complete mitochondrial genome of the brown pansy butterfly, Junonia stygia (Aurivillius, 1894), (Insecta: Lepidoptera: Nymphalidae). Mitochondr DNA B Resour. 5:41–43.
- Marcus JM. 2018. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genetics. 5(1):1–23,
- McCullagh BS, Marcus JM. 2015. The complete mitochondrional genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia-Pacific Ent. 18(4):749–755.
- Mutanen M, Wahlberg N, Kaila L. 2010. Comprehensive gene and taxon coverage elucidates radiation patterns in moths and butterflies. Proc Biol Sci. 277(1695):2839–2848.
- Park JS, Kim MJ, Jeong SY, Kim SS, Kim I. 2016. Complete mitochondrial genomes of two gelechioids, Mesophleps albilinella and Dichomeris ustalella (Lepidoptera: Gelechiidae), with a description of gene rearrangement in Lepidoptera. Curr Genet. 62(4):809–826.
- Payment JE, Marcus JM, Lalonde MML. 2020. The complete mitochondrial genome of the African leaf butterfly Kallimoides rumia (Insecta: Lepidoptera: Nymphalidae. Mitochondr DNA B Resour. 5(3):3415–3417.
- Quesada F. 2019. Área de Conservación Guanacaste, Fuente de Vida y Desarrollo: Macrosoma conifera (Heydilidae). [Accessed 28 July 2020]. https://www.acguanacaste.ac.cr/paginas-de-especies/insectos/268-hedylidae/540-i-macrosoma-conifera-i-hedylidae.
- Regier JC, Zwick A, Cummings MP, Kawahara AY, Cho S, Weller S, Roe A, Baixeras J, Brown JW, Parr C, et al. 2009. Toward reconstructing the evolution of advanced moths and butterflies (Lepidoptera: Ditrysia): an initial molecular study. BMC Evol Biol. 9(1):280.
- Scoble MJ. 1986. The structure and affinities of the Hedyloidea: a new concept of the butterflies. Bull Br Mus Nat Hist, Entomol Ser. 53:251–286.
- Scoble MJ, Aiello A. 1990. Moth-like butterflies (Hedylidae: Lepidoptera): a summary, with comments on the egg. J Nat Hist. 24(1):159–164.
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539.
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, Massachusetts, USA.
- Timmermans MJTN, Lees DC, Simonsen TJ. 2014. Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol. 79:169–178.
- Wahlberg N, Braby MF, Brower AVZ, de Jong R, Lee M-M, Nylin S, Pierce NE, Sperling FAH, Vila R, Warren AD, et al. 2005. Synergistic effects of combining morphological and molecular data in resolving the phylogeny of butterflies and skippers. Proc Biol Sci. 272(1572):1577–1586.
- Weintraub JD, Miller JS. 1987. The structure and affinities of the Hedyloidea: a new concept of butterflies. Cladistics. 3:299–304.
- Weller SJ, Pashley DP. 1995. In search of butterfly origins. Mol Phylogenet Evol. 4(3):235–246.
- Yack JE, Johnson SE, Brown SG, Warrant EJ. 2007. The eyes of Macrosoma sp. (Lepidoptera: Hedyloidea): A nocturnal butterfly with superposition optics. Arthropod Struct Dev. 36(1):11–22.
- Zhang J, Cong Q, Shen J, Brockmann E, Grishin NV. 2019. Three new subfamilies of skipper butterflies (Lepidoptera, Hesperiidae). Zookeys. 861:91–105.
