Abstract
The superfamily Gonodactyloidea is polyphyletic because of Hemisquillidae, but to date, mitochondrial genome of that family does not exist. As valuable data that can be compared in the future with that family within this superfamily, we report the first complete mitochondrial genome sequence of Taku spinosocarinatus of the family Takuidae. The mitochondrial genome is 15,960 bp in length and consists of 13 protein-coding genes (PCGs), 22 transfer RNA genes, 2 ribosomal RNA genes, and a non-coding A + T rich region. The overall base composition in the heavy strand was as follows: A: 34.2%, G: 13.4%, C: 19.8%, and T: 32.6%, with a G + C content of 33.2%. Phylogenetic analysis revealed that this species was most closely related to Gonodactylus chiragra of Gonodactylidae, registered with NCBI to date. The result of this study will enable additional comparisons between families in the future.
The crustacean order Stomatopoda consists of seven superfamilies and they have large and powerful raptorial appendages that can be used for ‘spearing’ or ‘smashing’ (Caldwell and Dingle Citation1976). Among them, Gonodactyloids comprise seven families and most have a smashing-type raptorial claw. According to stomatopod molecular phylogenetic studies, this superfamily is polyphyletic (Ahyong Citation1997; Hof Citation1998; Ahyong and Harling Citation2000; Wal et al. Citation2017) because of the family Hemisquillidae. The present species, Taku spinosocarinatus belongs to Takuidae within Gonodactyloidea has a smashing-type of raptorial claw like Hemisquillids. Since previous study (Wal et al. Citation2017) showed that smashing group in other superfamily had a monophyletic origin, the case of these families is an exception. Unfortunately, studies on complete mitochondrial genome have not progressed. Aiming to be able to comparable with the family Hemisquillidae and improve the phylogenetic knowledge on these families within this superfamily, and further characterize their mitochondrial gene order by sequencing the mitochondrial genome of Taku spinosocarinatus.
The specimen was collected by scuba diving in the subtidal zone of Dokdo Island, South Korea (geographic location: 37°14′34.9″N 131°52′08.6″E) on 1 June 2019, and was preserved in 95% ethyl alcohol. The specimen was deposited at the Research Institute of EcoScience, Ewha Womans University (EWNHMAR768). Total DNA was extracted from leg muscle tissue using DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) and the DNA library was prepared using TruseqNano DNA Prep Kit (Illumina, San Diego, CA, USA). The mitochondrial DNA was sequenced using Illumina Novaseq 6000 and MITObim (Hahn et al. Citation2013) was used for the assembly of the complete mitochondrial genome, which was annotated using MITOS (Bernt et al. Citation2013).
The mitochondrial genome comprised a total of 15,960 bp, which encoded 13 proteins, 22 transfer RNAs, two ribosomal RNAs, and a putative control region. For the protein-coding genes (PCGs), the most common shared start codon was ATG identified in COX2, COX3, NAD3, NAD4, NAD5, NAD4L, ATP6, and CYB. The starting codon used by COX1 was ACG, which is often observed in the COX1 gene of malacostracan mtDNAs (Cook Citation2005; Liu and Cui Citation2010), whereas for NAD1 was ATA, for ATP8 was ATC, and for NAD2 and NAD6 was ATT. The most frequent stop codon was TAA, except for COX2, NAD1, and NAD6 that had AAT, TAG, and CCT as stop codons. In particular, NAD1 and NAD6 proteins ended with an incomplete stop codon. Such cases have been identified in several PCGs of all stomatopod mitochondrial genomes published to date, which is attributed to excessive polyadenylation (Ojala et al. Citation1980, Citation1981). The overall mitochondrial base composition of this genome was A: 34.2%, T: 32.6%, G: 13.4%, and C: 19.8%, with a G + C content of 33.2%. The length of LrRNA and SrRNA genes in this species was 1367 and 837 bp, respectively., while the length of the transfer RNAs identified ranged from 65 to 74 nucleotides. The putative control region comprised 1069 bp and was located after transfer RNA-Val and SrRNA.
To explore the phylogenetic position of T. spinosocarinatus, we investigated the molecular phylogenetic relationships among stomatopod species using the complete mitochondrial genome sequence of seven species (). The phylogenetic tree was constructed based on sequences of 13 PCGs identified by the maximum likelihood (ML) method using MEGA X (Kumar et al. Citation2018). The GTR + G + I model was identified as the best-fit model for the data, using ModelFinder (Kalyaanamoorthy et al. Citation2017) with 1000 bootstrap replicates.
Figure 1. Phylogenetic tree of complete mitochondrial genomes from eight stomatopods (Oratosquilla oratoria (NC014342), Gonodactylus chiragra (NC007442), Harpiosquilla harpax (NC006916), Squilla empusa (NC007444), Squilla mantis (NC006081) Lysiosquillina maculata (NC007443), and Taku spinosocarinatus (MT672285)) constructed using maximum likelihood (ML) method.
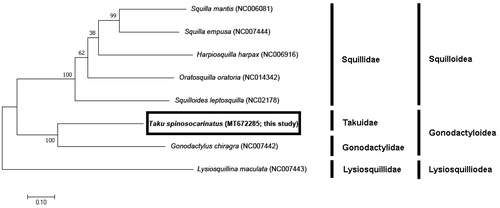
One gonodactyloid species and five squilloid species were used, with a lysiosquilloid species as an outgroup. Phylogenetic analysis revealed that this species was most closely related to Gonodactylus chiragra of Gonodactylidae, which is a smashing group within Gonodactyloidea and registered with NCBI to date. As this is the first record of the complete mitogenome sequence of the family Takuidae, it will enable further comparisons between families for future studies about this superfamily.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. MT672285. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA661984, SRR12640676, and SAMN16072594, respectively. The data that support the findings of this study are also openly available in Mendeley Data at http://dx.doi.org/10.17632/kg3ypr2cww.1
Additional information
Funding
References
- Ahyong ST. 1997. Phylogenetic analysis of the stomatopoda (Malacostraca). J Crustac Biol. 17(4):695–715.
- Ahyong ST, Harling C. 2000. The phylogeny of the stomatopod Crustacea. Aust J Zool. 48(6):607–642.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Caldwell RL, Dingle H. 1976. Stomatopods. Sci Am. 234(1):80–89.
- Cook CE. 2005. The complete mitochondrial genome of the stomatopod crustacean Squilla mantis. BMC Genomics. 6:105.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129
- Hof CHJ. 1998. Fossil stomatopods (Crustacea: Malacostraca) and their phylogenetic impact. J Nat Hist. 32(10–11):1567–1576.
- Kalyaanamoorthy S, Minh BQ, Wong TK, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Liu Y, Cui Z. 2010. The complete mitochondrial genome of the mantid shrimp Oratosquilla oratoria (Crustacea: Malacostraca: Stomatopoda): novel non-coding regions features and phylogenetic implications of the Stomatopoda. Comp Biochem Physiol Part D Genomics Proteomics. 5(3):190–198.
- Ojala D, Merkel C, Gelfand R, Attardi G. 1980. The tRNA genes punctuate the reading of genetic information in human mitochondrial DNA. Cell. 22(2 Pt 2):393–403.
- Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature. 290(5806):470–474.
- Wal CVD, Ahyong ST, Ho SYW, Lo N. 2017. The evolutionary history of Stomatopoda (Crustacea: Malacostraca) inferred from molecular data. Peer J. 5:e3844.
