Abstract
The Liu’s skink, Plestiodon liui, is endemic to China. In this study, a nearly complete mitochondrial genome (mitogenome) (17,643 bp in length) of P. liui from Junzhang Mountain, Wuxi, Jiangsu province, China, was determined by next-generation sequencing. Similar to the typical mtDNA of vertebrates, it contained two ribosomal RNA genes, 13 protein-coding genes (PCGs), 22 transfer RNA genes, and one control region (CR). With exception to the CR, all of the 37 genes were completely recovered. The PCGs were used to perform Bayesian phylogenetic analyses together with other scincid lizards with mitogenome data in GenBank. The resulting phylogenetic tree supported the monophyly of the P. capito group, and suggested that P. liui is the sister taxon to P. capito plus P. tunganus. The mitogenome of P. liui will provide fundamental data for the exploration of the mitogenome evolution in skinks.
In this study, we determined the mitochondrial genome (mitogenome) of the Liu’s skink, Plestiodon liui, a blue-tailed skink endemic to China (Hikida and Zhao Citation1989), by using the next-generation of sequencing, with GenBank accession number MT662111. The specimen (voucher number CB202004001) was collected from Junzhang Mountain (31.442742˚N, 120.223861˚E), Wuxi city, Jiangsu province, China on 1 April 2020. Its liver tissue was fixed with 95% ethanol, and stored at −20 °C in the herpetological collection, Chengdu Institute of Biology, Chinese Academy of Sciences. A small amount of liver tissue was shipped to Sangon Bitotech (Shanghai, China) for genomic extraction and 150-base-pair paired-end library construction; sequencing was performed on an Illumina Hiseq 2000 instrument (Illumina, San Diego, CA). De novo assembly of clean reads was performed using SPAdes v3.11.0 (Bankevich et al. Citation2012). The mitogenome of Pleistiodon tunganus (MK370739; Chen et al. Citation2019) was further used as a reference to assemble that of P. liui. The sequence was annotated with the MITOS (Bernt et al. Citation2013) webserver; meanwhile, the complete mitogenomes of some other skink species available in GenBank were also used as reference points for annotations.
The nearly complete mitogenome of P. liui is 17,643 bp in length, which is composed of 24.7% (T), 29.6% (C), 30.5% (A), 15.2% (G). The mitogenome of P. liui is similar to the typical vertebrate mitochondrial genome. It encodes 37 genes, containing 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (12S rRNA and 16S rRNA), 13 protein-coding genes (PCGs), and a control region (CR or D-loop). In the nearly complete mitogenome of P. liui, most of the genes are distributed on the H- strand, except for the ND6 subunit gene and 8 tRNA genes (tRNA-Gln, Ala, Asn, Cys, Tyr, Ser[UCN], Glu, and Pro), which are distributed on the L-strand. In the 13 PCGs, 12 PCGs are initiated with the typical ATG codon, only COX1 with GTG; three PCGs (ND1, ND2, Cytb) use TAG as stop codon; four PCGs (ATP8, ATP6, ND4L, ND5) use TAA as stop codon; COX1 and ND6 use AGA and AGG as stop codons, respectively; the others (COX2, COX3, ND3, ND4) terminate with T as an incomplete stop codon. As for the CR, 375 bp were already determined adjacent to tRNA-Pro, along with 1851 bp prior to tRNA-Phe.
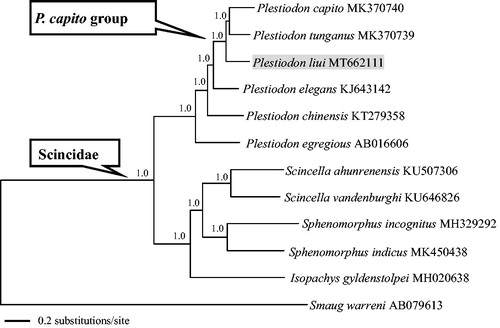
The concatenated PCGs of Scincidae available in GenBank and Smaug warren (Cordylidae) as outgroup were used to reconstruct the Bayesian phylogenetic tree for assessing mitochondrial sequence authenticity of P. liui and its phylogenetic placement. DNA sequences were aligned in MAFFT v7.313 (Katoh and Standley Citation2013). The PCGs were translated into amino acid sequences, and then all sequences were concatenated into a nucleotide data set (11,388 bp in total) through PhyloSuite v1.2.1 (Zhang et al. Citation2020). The plug-in programs in PhyloSuite were used for gene partitioning, model selection, and tree reconstruction. The best-fitting substitution models and partitioning schemes were selected in PartitionFinder v2.1.1 using the Bayesian information criterion (Lanfear et al. Citation2017). Partitioned Bayesian analyses were conducted using MrBayes v.3.2.6 (Ronquist et al. Citation2012). As shown in , the phylogenetic tree recovered the monophyly of Scincidae and the P. capito species group (Brandley et al. Citation2011, Citation2012), and suggested that P. liui is the sister taxon to P. capito plus P. tunganus (Cai et al. Citation2019). The mitogenome of P. liui will provide fundamental data for the exploration of the mitogenome evolution in skinks (Scincidae).
Figure 1. A majority-rule consensus tree inferred from Bayesian inference using MrBayes with the best models for each partition, based on the PCGs of 11 species of Scincidae and one outgroup. The phylogenetic placement of P. liui is highlighted. GenBank accession numbers are given with species names. Node numbers show Bayesian posterior probabilities. Branch lengths represent means of the posterior distribution.
Acknowledgments
We thank Dr. Weizhao Yang (Lund University) for helping to assemble the mitochondrial genome, and Mr. Minggang Xu for collecting the specimen of Plestiodon liui. We extend our thanks to an anonymous Associate Editor for his/her critical review.
Disclosure statement
No potential conflict of interest was reported by the authors. The authors alone are responsible for the content and writing of this article.
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov/, reference number [MT662111], or available from the corresponding author.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin V, Nikolenko S, Prjibelski A, Pyshkin A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Brandley MC, Ota H, Hikida T, de Oca ANM, Ferıa-Ortız M, Guo X, Wang Y. 2012. The phylogenetic systematics of blue-tailed skinks (Plestiodon) and the family Scincidae. Zool J Linn Soc. 165(1):163–189.
- Brandley MC, Wang Y, Guo X, de Oca ANM, Ferıa-Ortız M, Hikida T, Ota H. 2011. Accommodating heterogenous rates of evolution in molecular divergence dating methods: an example using intercontinental dispersal of Plestiodon (Eumeces) lizards. Syst Biol. 60(1):3–15.
- Cai B, Yao M, Guo X. 2019. First record of Plestiodon liui from Henan province, China and analysis of its phylogenetic placement. Chin J Zool. 54:668–677.
- Chen M, Liu J, Chen D, Guo X. 2019. The complete mitochondrial genome of a blue-tailed skink (Plestiodon tunganus) endemic to Sichuan Basin. Mitochondrial DNA B. 4(1):1109–1110.
- Hikida T, Zhao E. 1989. Eumeces liui: a new species of blue-tailed skink (Lacertilia: Scincidae) from China. Copeia. 1989(1):110–114.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Ronquist F, Teslenko M, Mark PVD, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Zhang D, Gao F, Li WX, Jakovlić I, Zou H, Zhang J, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
