Abstract
In this paper, we determined and described the complete mitochondrial genome of Robin Accentor (Prunella rubeculoides), the first complete mitogenome. The complete genome of P. rubeculoides was 16,796 bp in length and contained 13 protein-coding genes, 22 transfer RNA genes, two ribosome RNA genes, and one non-coding control region. The overall base composition of the mitochondrial DNA was 29.7% for A, 23.7% for T, 15.6% for G, and 31.0% for C, with a GC content of 46.6%. This information of P. rubeculoides mitogenome is significance for phylogenetic studies of the family Prunellidea.
Robin Accentor (Prunella rubeculoides) is a grayish and medium-size bird (16 cm) within the family Prunellidea. Its chest is chestnut brown, while its head, throat, upper body, wings, and tail are smoke-brown. Its upper back has a faint black stripe. It has a narrow black collar between its gray throat and chestnut-brown chest. The rest of its body is white (Birdnet Citation2014). The bird distributes in Bhutan, China, India, Nepal, and Pakistan, which is completely endemic to the Palearctic (Surhone et al Citation2011). The species was rated in the least concerned species and placed on the IUCN's Citation2016 Red List of Least Concern Species (LC) (IUCN Citation2016). In China, it is found in the Himalayas, central China and south of the Qinghai-Tibet Plateau (Surhone et al. Citation2011). It is an uncommon resident bird, found in the meadows and rhododendron and willow thicles of northern and eastern Qinghai, western Gansu and Sichuan, and southern Tibet, 3600–4900 m above sea level (Birdnet Citation2014). However, there is no molecular data reported on P. rubeculoides. Therefore, in order to better understand the phylogeny and evolution of this species or subspecies in the Prunellidea family, we sequenced the complete mitochondrial genome of P. rubeculoides.
A natural dead P. rubeculoides was collected from Xiejia village, Marong township, Baiyu county, Ganzi Tibetan Autonomous region, Sichuan province. The specimens is now in the Key Laboratory of Bioresources and Ecoenvironment, Sichuan University. The stored number of the sample is 2019PD-1. Total genomic DNA was extracted from muscle tissue using standard phenol-chloroform methods (Sambrook and Russell Citation2001). The complete mitochondrial genome was amplified using PCR and PCR produces were sequenced by ABI PRISM 3730 DNA sequencer. The complete mitochondrial genome of P. rubeculoides was submitted to the NCBI database under the accession number QYL.sqn DG2 MT903225. The full genome of P. rubeculoides mitochondrial is a circular DNA molecule and is 16,796 bp in length. The overall base composition was 29.7% A, 23.7% T, 15.6% C, and 31.0% G, with a GC ratio of 46.6%. The complete mitogenome of P. rubeculoides included 13 protein-coding genes, two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA (tRNA) genes, and one non-coding control region (D-loop). Among these genes, ND6 and eight tRNA genes (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu, and tRNA-Pro) located on the light strand (L-strand), whereas other genes located on the heavy strand (H-strand). The 12S and 16S rRNA genes located between tRNA-Phe and tRNA-Leu contains 974 and 1596 bp, respectively.
Eleven of the 13 PCGs used ATG as the start codon, while COX1 and ND6 selected GTG and CTA, respectively. The PCGs have six types of termination codon, including TAA for ATP8, ND3, ND4 and Cytb, TAG for ATP6, AGA for ND1 and ND5, AGG for COX1, ‘T––’ for COX2, COX3, ND4 and ND6, and ‘TA–’ for ND2. Some birds have been found to have single insertion mutations in ND3, but the P. rubeculoides has WOC (without extra Cytosine) ND3 (Yan et al. Citation2017). Unlike these mitochondrial genomes of many other bird, which have a pseudo—control region (Song et al. Citation2015), the mitochondrial genome of P. rubeculoides has only one single control region, and no repeat sequence was found in the control region.
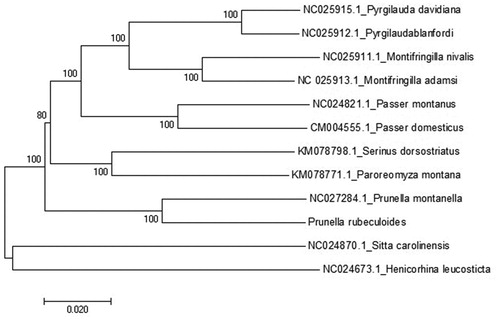
A neighbor-joining (NJ) tree of 11 species from Passeriformes was constructed based on the dataset of 13 concatenated mitochondrial PCGs using MEGA 7 (MEGA Inc., Englewood, NJ) with 1000 bootstrap replicates. The reconstructed phylogenetic tree supports the position of P. rubeculoides in the Prunellidea () with high bootstrap values. As can be seen from the tree (), the results are similar to previous studies on Prunella montanella in the same genus (Yao et al. Citation2015). The sequence data will be useful for the phylogenetic studies of the family Prunellidea species in the future.
Disclosure statement
The authors report no conflicts of interest and alone are responsible for the content and writing of the article.
Data availability statement
We also confirm that the data supporting the findings of this study are available within the article and in Genbank. https://www.ncbi.nlm.nih.gov/Genbank/update.html
Additional information
Funding
References
- Birdnet. 2014. Robin Accentor. http://www.cnniao.com/niao/1432.html.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor: Cold Spring Habor Lab Press; p. A8.40–A8.51.
- Song XH, Jie Huang J, Yan CC, Xu GW, Zhang XY, Yue BS. 2015. The complete mitochondrial genome of Accipiter virgatus and evolutionary history of the pseudo-control regions in Falconiformes. Biochem Syst Ecol. 58:75–84.
- Surhone LM, Tennoe MT, Henssonow SF. 2011. Robin Accentor [M]. Betascript Publishing.
- IUCN. 2016. The IUCN Red List of Threatened Species. e.T22718624A94589670. https://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T22718624A94589670.en.
- Yan C, Mou B, Meng Y, Tu F, Fan Z, Price M, Yue BS, Zhang XY. 2017. A novel mitochondrial genome of Arborophila and new insight into Arborophila evolutionary history. PLoS One. 12(7):e0181649
- Yao J, Zhao X, Li Y, Li L, Yan S. 2015. The complete sequence of mitochondrial genome of Siberian accentor (Prunella montanella). Mitochondrial DNA. 27(5):1–2.