Abstract
We sequenced and annotated mitochondrial genome (mitogenome) of oilseed rape pest Psylliodes punctifrons Baly for the first time. The mitogenome is 15,611 bp, and the nucleotide composition of 37 genes is highly A + T biased (A: 34.2, C: 11.6, G: 12.1, and T:42.1). All PCGs start with ATN and stop with TAR, except COX3 and ND4 that stop with incomplete codon T—. The phylogenetic tree confirms that P. punctifrons is clustered with other Psylliodes species. This study enriches the mitogenomes of agricultural pests.
The genus of Psylliodes Latreille belongs to the subfamily Galerucinae and includes more than 200 species (He et al. Citation2019). A lot of species within this genus are known as agricultural pests, which can attach plants of some 30 families (Jolivet and Hawkeswood Citation1995). Psylliodes punctifrons is widely distributed throughout China and is pest of all cruciferous vegetables (He et al. Citation2000). In this study, we have sequenced and determined the mitochondrial genome (mitogenome) of P. punctifrons using next-generation sequencing method (Illumina) for the first time. The sequences obtained in this study may facilitate future studies on identification, population genetics, and biological control of P. punctifrons.
Total genome DNA of P. punctifrons was extracted from male adults that were collected from eggplants, in Guiyang, Guizhou Province, China (106°39′59″E, 26°30′20″N), in June 2020. Total genomic DNA was extracted using DNeasy Blood and Tissue Kit (Qiagen), following the protocol. Paired-end library (450 bp) was sequenced using Illumina Hiseq4000 platform, with 150 bp pair-end sequencing method (Sangon Biotech (Shanghai) Co., Ltd.), after trimming and filtering of the raw sequencing reads, the Q20 bases ratio is 97.00%, generating 24,477,104 high-quality clean reads (7.47 Gb). Remaining samples and genome DNA are deposited in the Institute of Entomology, Guizhou University, Guiyang, China (GUGC).
Mitogenome of P. punctifrons was assembled using Geneious primer (v. 2019.2.1) with reference sequences (Psylliodes hispanus, KX943503), Assembled mitogenome of P. punctifrons is 15,611 bp in length (GenBank No. MT890591), and 39,065 reads were matched to the reference sequences, the mean coverage of the mitogenome is 354.5. Mitogenome of P. punctifrons were annotated using MITOS v2 server with the invertebrate genetic codes (Bernt et al. Citation2013), ARWEN 1.2 (Laslett and Canback Citation2008) and ORF Finder in NCBI. containing typical 37 mitogenome genes (13 protein-coding genes (PCGs, 11,129 bp), 22 transfer RNA genes (tRNAs, 1,447 bp), and two ribosomal RNA genes (rRNAs, 2021 bp)) and a control region. Generally, the nucleotide composition of 37 genes is highly A + T biased (A: 34.2, C: 11.6, G: 12.1, and T:42.1), except COX3 and ND4 stop with incomplete codon T—, all PCGs start with the canonical putative start codon ATN (ATA/ATT/ATC/ATG), and stop with termination codon TAA/TAG. The 16S rRNA and 12S rRNA are 1280 bp and 741 bp in length, respectively.
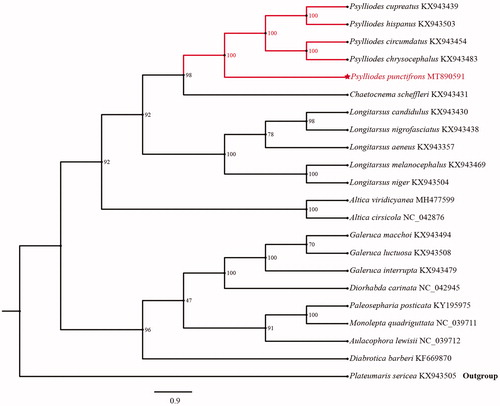
The phylogenetic relationships of P. punctifrons were reconstructed with IQ-TREE using an ultrafast bootstrap that approximation approach1000 replicates based on first and second codon positions of the 13 PCGs with 7234 sites. Each PCG sequence was aligned using MAFFT v7.0 and Gblocks 9.1 b (Katoh and Standley Citation2013) and then concatenated individual genes using MEGAX (Kumar et al. Citation2018). The phylogenetic tree confirms that P. punctifrons is clustered with other Psylliodes species with high support (100) (). We hope that our data can be useful for further study.
Figure 1. Phylogenetic analyses of Psylliodes punctifrons based upon first and second codon positions of the 13 PCGs of 22 species. This analysis was performed using IQ-TREE software. Numbers at nodes are bootstrap values. The accession number for each species is indicated after the scientific name.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number MT890591.
References
- He C, Pan F, Wang G, Pei X. 2000. Damage grades and habits of Psylliodes punctifrons Baly. J Gansu Agric Univ. 35(4):377–381.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Laslett D, Canback B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Jolivet P, Hawkeswood TJ. 1995. Host plants of Chrysomelidae of the world. London: Backhuys Publ; p. 281.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- He Y, Zeng H, Fan Y, Ji S, Wu J. 2019. Application of deep learning in integrated pest management: a real-time system for detection and diagnosis of oilseed rape pests. Mobile Information Syst. 2019:1–14.
