Abstract
We completed chloroplast genome of Douinia plicata (Lindb.) Konstant. & Vilnet., presenting morphological features including denticulate leaf margin, verrucose cuticle on base of leaves, and 80–100° keel angle with stem at the midleaf. It is 118,797 bp long (GC ratio is 33.9%) and has four subregions: 81,142 bp of large single copy (31.9%) and 19,611 bp of small single copy (31.0%) regions are separated by 9,017 bp of inverted repeat (46.3%) regions including 130 genes (86 protein-coding genes, eight rRNAs, and 36 tRNAs). Phylogenetic trees show D. plicata is clustered with two Scapania species.
Genus Douinia (C.E.O. Jensen) H. Buch is a small genus of leafy liverworts covering three accepted species (Söderström et al. Citation2016). According to recent molecular phylogenetic studies of genus Douinia, Macrodiplophyllum plicatum (Lindb.) Perss. was transferred to Douinia from Macrodiplophyllum (H. Buch) Perss. (Vilnet et al. Citation2010; Konstantinova et al. Citation2013). Douinia plicata (Lindb.) Konstant. & Vilnet., the arctic-boreal, amphipacific species in Douinia genus, is distributed in the Northeast Asia (Korea, Japan, China, and Russian Far East; Bakalin Citation2010; Choi et al. Citation2012). D. plicata is distinct from the other Douinia species due to entire to denticulate leaf margin, striolate to verrucose cuticle on base of leaves, 80–100° keel angle with stem at the midleaf (Choi et al. Citation2012). Moreover, it is endemic to Northeast Asia, which can be a good candidate for understanding genetic features of Scapaniaceae in the world. Here, we reported completed chloroplast genome sequences of D. plicata.
The plants of D. plicata collected in Taebaek city, Korea (Voucher in Jeonbuk National University Herbarium (JNU); 5 Oct. 2019, S.S. Choi, CS-1910996b; 37.101486 N, 128.917547E) was used for extracting DNA with DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using NovaSeq6000 at Macrogen Inc., Korea, acquiring 7.5 million 151-bp pair-end raw reads (2.28 Gbp). Chloroplast genome was completed by Velvet v1.2.10 (Zerbino and Birney Citation2008), SOAPGapCloser v1.12 (Zhao et al. Citation2011), BWA v0.7.17 (Li Citation2013), and SAMtools v1.9 (Li et al. Citation2009) under the environment of Genome Information System (GeIS; http://geis.infoboss.co.kr/; Park et al., in preparation). Geneious R11 11.0.5 (Biomatters Ltd, Auckland, New Zealand) was used for annotation based on S. amplicata chloroplast genome (MT644123; Choi et al. Citation2020).
The chloroplast genome of D. plicata (GenBank accession is MT898431) is 1,18,797 bp long (GC ratio is 33.9%) and has four subregions: 81,142 bp of large single copy (31.9%) and 19,611 bp of small single copy (31.0%) regions are separated by 9,017 bp of inverted repeat (IR; 46.3%). It contained 130 genes (86 protein-coding genes, eight rRNAs, and 36 tRNAs); nine genes (four rRNAs and five tRNAs) are duplicated in IR regions. Gene order of D. plicata chloroplast is identical to those of Scapania species.
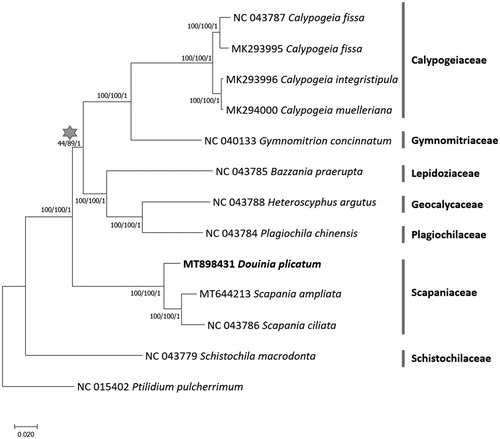
Thirteen complete chloroplast genomes including D. plicata were used for constructing neighbor-joining (bootstrap repeat is 10,000), maximum likelihood (bootstrap repeat is 1000), and Bayesian Inference phylogenic trees using MEGA X (Kumar et al. Citation2018) and MrBayes v3.2.7a (Huelsenbeck and Ronquist Citation2001) after aligning whole chloroplast genome sequences using MAFFT v7.450 (Katoh and Standley Citation2013). Phylogenetic trees show that D. plicata is clustered with two Scapania chloroplast genomes that belong to the same family, Scapaniaceae, with high supportive values (). It is congruent with previous phylogenetic study (Heinrichs et al. Citation2012). In addition, Scapaniaceae clade is located outside of the rest families except Schistochilaceae family, which is different from the previous study (Choi et al. Citation2020), displaying relatively low supportive values (; see gray star). It requires more chloroplast genomes of Jungermanniales species. Our chloroplast genome together with up-coming additional chloroplast genome of Jungermanniales will provide their detailed phylogenetic relationship.
Figure 1. Neighbor joining (bootstrap repeat is 10,000), maximum likelihood (bootstrap repeat is 1000), and Bayesian Inference phylogenetic trees of twelve complete chloroplast genomes: Douinia plicata (MT898431 in this study), Scapania ampliata (MT644123; Choi et al. Citation2020), Scapania ciliata (NC_043786), Bazzania praerupta (NC_043785), Calypogeia fissa (NC_043787), Gymnomitrion concinnatum (NC_040133; Myszczyński et al. Citation2018), Heteroscyphus argutus (NC_043788), Plagiochila chinensis (NC_043784), Schistochila macrodonta (NC_043779), Calypogeia fissa (MK293995), Calypogeia integristipula (MK293996), Calypogeia muelleriana (MK294000), and Ptilidium pulcherrimum (NC_015402; Forrest et al. Citation2011) as an outgroup. Gray-filled start indicates the clade displaying low supportive values. Phylogenetic tree was drawn based on the maximum likelihood phylogenetic tree. The numbers above branches indicate bootstrap support values of maximum likelihood, neighbor joining, and Bayesian Inference phylogenetic trees, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Chloroplast genome sequence can be accessed via accession number MT898431 in NCBI GenBank.
Additional information
Funding
References
- Bakalin VA. 2010. Hepaticae of the Kuril Islands (northwestern Pacific): a transoceanic route from circumboreal to East Asian flora. Ann Bot Fennici. 47(2):81–105.
- Choi SS, Bakalin VA, Sun B-Y. 2012. Scapania and Macrodiplophyllum in the Russian Far East. Bot Pac. 01 (1):31–95.
- Choi SS, Kwon W, Park J. 2020. The complete chloroplast genome of Scapania ampliata Steph., 1897 (Scapaniaceae, Jungermanniales). Mitochondrial DNA B. 5(3):2908–2910.
- Forrest LL, Wickett NJ, Cox CJ, Goffinet B. 2011. Deep sequencing of Ptilidium (Ptilidiaceae) suggests evolutionary stasis in liverwort plastid genome structure. Plecevo. 144(1):29–43.
- Heinrichs J, Bombosch A, Feldberg K, Kreier H-P, Hentschel J, Eckstein J, Long D, Zhu R-L, Schäfer-Verwimp A, Schmidt AR, et al. 2012. A phylogeny of the northern temperate leafy liverwort genus Scapania (Scapaniaceae, Jungermanniales). Mol Phylogenet Evol. 62(3):973–985.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Konstantinova NA, Vilnet AA, Söderström L, Hagborg A, Von Konrat M. 2013. Notes on early land plants today. 14. Transfer of two Macrodiplophyllum species to Douinia (Scapaniaceae, Marchantiophyta). Phytotaxa. 76(3):31–32.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303.3997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Myszczyński K, Górski P, Ślipiko M, Sawicki J. 2018. Sequencing of organellar genomes of Gymnomitrion concinnatum (Jungermanniales) revealed the first exception in the structure and gene order of evolutionary stable liverworts mitogenomes. BMC Plant Biol. 18(1):1–12.
- Söderström L, Hagborg A, Von Konrat M. 2016. Early land plants today: index of liverworts & hornworts 2013–2014. Phytotaxa. 269(3):133–185.
- Vilnet A, Konstantinova N, Troitsky A. 2010. Molecular insight on phylogeny and systematics of the Lophoziaceae, Scapaniaceae, Gymnomitriaceae and Jungermanniaceae. Arctoa J Bryol. 19(1):31–50.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
