Abstract
Euphorbia lathyris L. is a well-known bioenergy plant cultivated in many parts of the world. In this study, we sequenced the complete chloroplast (cp) genome sequence of E. lathyris to investigate its phylogenetic relationship in the family Euphorbiaceae. The cp genome was 163,738 bp in length, consisting of a pair of inverted repeats (IRa and IRb: 26,837 bp) separated by a large single-copy region (LSC: 91,783 bp) and a small single-copy region (SSC: 18,281 bp). The GC content of whole cp genome is 35.6%. Annotation showed the presence of 113 unique genes with 79 protein-coding genes, four tRNA genes, and 30 rRNA genes. Phylogenetic analysis indicated that E. lathyris was in the basal position of subgen. Esula, closely related to sect. Esula and sect. Helioscopiae.
Euphorbia lathyris L., or Caper spurge, is a very isolated species that is adventive in many parts of the world. It is probably native only in the Mediterranean region (Wu et al. Citation1994). The species is cultivated as an ornamental plant or developed as a commercial crop (Govaerts et al. Citation2000). With the gradual depletion of nonrenewable fossil energy and the rising oil price, people pay more attention to the development of bioenergy. E. lathyris is a kind of energy plant which can produce diesel oil. The oil content of its seeds is generally about 45%. All parts of the plant, including the seeds and roots, are poisonous. The seeds are used medicinally as a violent purgative in folk medicine.
Total genomic DNA from fresh leaves was extracted by using the method of Li et al. (Citation2013). Voucher specimen of E. lathyris were collected from Li county, Hunan province, China(29°37′48″N, 111°45′36″E) and was stored at the herbarium of Institute of Chinese Materia Medica (CMMI), China Academy of Chinese Medical Sciences with the voucher number is 430723LY1047. The sequencing library was constructed and quantified following the methods introduced by Dong et al. (Dong et al. Citation2017; Sun et al. Citation2020). The whole genome sequencing was conducted with 150 bp paired-end reads on the Illumina HiSeq X Ten platform. Contigs were assembled from the high-quality paired-end reads by using the SPAdes 3.6.1 program (Kmer = 95) (Bankevich et al. Citation2012). The chloroplast genome contigs selected by the Blast program (Altschul et al. Citation1990), taken Euphorbia esula (GenBank: KY000001) as the reference. The selected contigs were assembled using Sequencher 4.10. Gene annotation of E. lathyris was performed using DOGMA annotation (Wyman et al. Citation2004) and manually corrected for codons and gene boundaries using BLAST searches. The annotated cp genome of E. lathyris was submitted to the GenBank under the accession number MT830859.
The circular cpDNA of E. lathyris was 163,738 bp in length, consisting of a pair of inverted repeats (IRa and IRb: 26,837 bp) separated by a large single-copy region (LSC: 91,783 bp) and a small single-copy region (SSC: 18,281 bp). The GC content of whole cp genome is 35.6%. The cpDNA of E. lathyris comprised 113 distinct genes, including 79 protein-coding genes, four ribosomal RNA genes, and 30 transfer RNA genes. In these genes, 19 were duplicated in the IR regions and 19 genes contained one or two introns, 17 harbored a single intron, and two (ycf3,clpP) contained double introns.
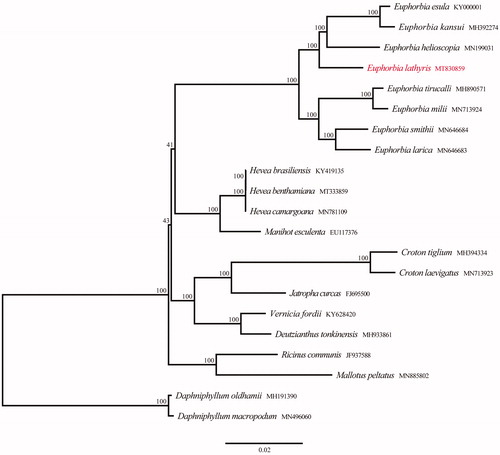
To investigate the phylogenetic relationships between E. lathyris and other related species in family Euphorbiaceae, Twenty chloroplast genome sequences were downloaded from GenBank to construct a phylogenetic IQ-tree using PhyloSuite under the TVM + F + I + G4 model with 1000 bootstrap replicates (Nguyen et al. Citation2015; Zhang et al. Citation2020). Daphniphyllum oldhamii and D. macropodum were taken as outgroups. The phylogenetic analysis revealed that samples of genus Euphorbia were strongly supported as monophyletic, E. lathyris was in the basal position of subgen. Esula additionally (). The cpDNA of E. lathyris is closly related to sect. Esula and sect. Helioscopiae. The complete chloroplast genome reported in this study will be a valuable resource for future studies on genetic diversity, taxonomy, and phylogeny of family Euphorbiaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI https://www.ncbi.nlm.nih.gov/, reference number MT830859, raw data BioProject ID: PRJNA660005, Biosample (SAMN15932209).
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Dong W, Xu C, Li W, Xie X, Lu Y, Liu Y, Jin X, Suo Z. 2017. Phylogenetic resolution in juglans based on complete chloroplast genomes and nuclear DNA sequences. Front Plant Sci. 8:1148.
- Govaerts R, Frodin DG, Radcliffesmith A. 2000. World checklist and bibliography of Euphorbiaceae (and Pandaceae). Kew: Royal Botanic Gardens.
- Li JL, Wang S, Jing Y, Wang L, Zhou SL. 2013. A modified CTAB protocol for plant DNA extraction. Chinese Bull Bot. 48:72–78.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Sun J, Wang Y, Liu Y, Xu C, Yuan Q, Guo L, Huang L. 2020. Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Sci Rep. 10(1):11466.
- Wu ZY, Raven PH, Hong DY. 1994. Flora of China. Vol. 44. Beijing: Science Press.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhang D, Gao F, Jakovlic I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.