Abstract
The complete mitochondrial genome (mitogenome) of Tropidophorus hangnam was sequenced from its paratype (GenBank accession no. MN977920). It was 16,777 bp in length with a base composition of 31.99% A, 29.49% C, 14.34% G, and 24.18% T, and a GC content of 43.83%. The genome includes 13 protein-coding genes (PCGs), 22 transfer RNA genes, 2 ribosomal RNA genes, and a control region (D loop). Most T. hangnam genes are located on the H strand, except for the ND6 gene and eight tRNA genes, which are located on the L strand. Phylogenetic analyses based on 13 PCGs indicated that T. hangnam is sister to the clade composed of the genera Scincella and Sphenomorphus. The newly sequenced T. hangnam mitogenome will provide basic data for further studies on the genetic diversity and molecular phylogenetic relationships of the genus Tropidophorus.
Chuaynkern et al. (Citation2005) first described Tropidophorus hangnam as a novel species. The holotype (THNHM 05776) and paratypes (THNHM 05777–85) were collected from the Phu Khaeo Wildlife Sanctuary, Chaiyaphum Province, northeastern Thailand. Tropidophorus hangnam is an endemic species known only from its type locality, with no additional specimens reported (Das Citation2010; Chanard et al. Citation2015); therefore, it is classified as data deficient (DD) on the International Union for Conservation of Nature (IUCN) Red List (Panitvong et al. Citation2018). Prior to the current work, no complete mitochondrial genome from the genus Tropidophorus or DNA sequences of T. hangnam were published in GenBank (https://www.ncbi.nlm.nih.gov/), and its genetic relationships with other genera of Scincidae based on mitogenome sequences are unknown. Therefore, we assembled and characterized the complete mitogenome of T. hangnam and clarified its relationships with other Scincidae genera using 13 protein-coding genes (PCG) sequences.
The paratype specimen (field no. 20502) was collected from the Phu Khaeo Wildlife Sanctuary, Chaiyaphum Province, Thailand (16°19′29′′ N 101°31′21′′ E), on 7 November 2004. Its liver tissue was fixed in 95% ethanol and stored at −20 °C. Genomic DNA was extracted from the liver tissue as described by Hillis et al. (Citation1996), with modifications. To amplify the T. hangnam mitochondrial genome, we designed specific polymerase chain reaction (PCR) primers based on mitochondrial genome sequences obtained from Plestiodon elegans (KJ643142), Scincella vandenburghi (KU646826), and Sphenomorphus incognitus (MH329292). PCR primers and protocols are available upon request. The DNA sequences were assembled using the AutoAssembler version 2.1.1 software (Applied Biosystems, Foster City, CA). The assembled mitogenome was annotated using the MITOS WebServer (Bernt et al. Citation2013), Dual Organellar Genome Annotator (Wyman et al. Citation2004), and MitoFish programs (Iwasaki et al. Citation2013). All transfer RNA (tRNA) genes were further confirmed using the tRNAscan-SE search server (Lowe and Chan Citation2016).
The complete T. hangnam mitogenome was 16,777 bp in length and was deposited in GenBank (accession no. MN977920). The genomes consisted of 13 PCGs, 22 tRNA genes, 2 ribosomal RNA (rRNA) genes, and a control region (D loop). The overall base composition of the heavy strand is 31.99% A, 29.49% C, 14.34% G, and 24.18% T, with a GC content of 43.83%. Most mitochondrial genes are encoded on the heavy strand, except for ND6 and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer, tRNAGlu, and tRNAPro) that are encoded on the light strand. Most of the 13 PCGs begin with the common initiation codon ATG, with only the COI gene beginning with GTG. Nine genes (ND1, ND2, COI, ATPase 8, ATPase 6, ND3, ND4L, ND5, ND6, and Cytb) end with complete stop codons (TAA, TAG, AGA, and AGG), and the other four genes (COII, COIII, ND3, and ND4) end with T as an incomplete stop codon, which is presumably completed as TAA by post-transcriptional polyadenylation (Anderson et al. Citation1981). The lengths of the 12S rRNA and 16S rRNA genes are 966 and 1519 bp, respectively. Two ribosomal subunit genes were separated by tRNAVal. The lengths of the 22 tRNA genes ranged from 65 bp (tRNACys) to 76 bp (tRNALeu), and the inferred secondary structures of all tRNAs conform to the characteristic structural features of mitochondrial tRNAs. The control region (D loop) is 1380 bp in length and located between the tRNAPro and tRNAPhe genes.
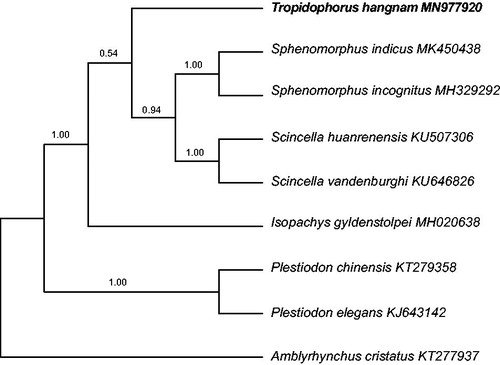
To explore the evolutionary placement of the genus Tropidophorus within the family Scincidae, we performed phylogenetic analyses using data from 13 PCGs of the mitogenomes of T. hangnam and 7 other skink species, with Amblyrhynchus cristatus (Iguanidae) as an outgroup. Phylogenetic analyses included both Bayesian inference (BI) and maximum parsimony (MP) analyses. BI was performed using the MrBayes version 3.2 software (Ronquist et al. Citation2012), and MP analyses were performed using the PAUP*software (Swofford Citation2003). Tree topologies obtained through BI and MP analyses based on the 13 PCGs were identical and statistically supported by high bootstrap and posterior probability values (). These results indicate that T. hangnam is closer to the clade comprising Scincella and Sphenomorphus than to that comprising the genera Isopachys and Plestiodon.
Acknowledgments
We thank the Department of Forest Biology, Faculty of Forestry, Kasetsart University, for providing necessary molecular facilities and support for this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data supporting the findings of this study are openly available from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) under reference no. MN977920.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290(5806):457–465.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chanard T, Parr JWK, Nabhitabhata JA. 2015. Field guide to the reptiles of Thailand. Oxford: Oxford University Press.
- Chuaynkern Y, Nabhitabhata J, Inthara C, Kamsook M, Somsri K. 2005. A new species of the water skink Tropidophorus (Reptilia: Squamata: Scincidae) from northeastern Thailand. Thailand Nat Hist Mus J. 1(2):165–176.
- Das I. 2010. A field guide to the reptiles of South-east Asia. London: New Holland Publishers.
- Hillis DM, Mable BK, Larson A, Davis SK, Zimmer EA. 1996. Nucleic acids IV: sequencing and cloning. In: Hillis DM, Moritz C, Mable BK, editors. Molecular systematics. 2nd ed. Sunderland (MA): Sinauer Associates; p. 321–384.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–57.
- Panitvong N, Cota M, Sumontha M. 2018. Tropidophorus hangnam. The IUCN red list of threatened species. IUCN red list threat species. [accessed 2020 July 13]. http://dx.doi.org/10.2305/IUCN.UK.2018-2.RLTS.T104832271A104853855.en.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland (MA): Sinauer Associates.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.