Abstract
Rheum likiangense Samuelsson (Polygonaceae) is an endangered alpine rhubarb in the Qinghai-Tibet Plateau. In this study we report the complete chloroplast genome sequence (plastome) of Rh. likiangense. The assembled plastome is 162,291 bp in length with 31,741 bp inverted repeat (IR) regions and 128 annotated genes, including 34 tRNA genes, 8 rRNA genes, and 86 protein-coding genes. Phylogenetic analyses based on the full plastome sequences suggest the close relationship of Rh. likiangense with Rh. acuminatum and Rh. nobile. The plastome reported here is highly useful for designing plastome SSR markers to conduct a further conservation genetic study of this endangered rhubarb.
Rheum likiangense Samuelsson (Polygonaceae) is distributed in the Qinghai-Tibet Plateau with altitudes from 3500 m to 4400 m and a well adaptation to the alpine habitat (Xie Citation2000). It is used as an important Tibetan medicine and the wild resources decreases rapidly in the recent past. It is now listed as a locally endangered species. An effective conservation strategy is needed to protect this rhubarb species. In this study, we sequenced and reported the complete chloroplast genome (plastome) of Rh. likiangense (GenBank: MT806193). Such a maternally inherited plastome sequence is critical for designing SSR markers to conduct population genetics study of this species. We further performed phylogenetic analyses of this species and other rhubarb species with plastomes available.
We collected fresh leaves of Rh. likiangense from Chengduo, Qinghai (33°10′56″N, 97°23′21″E, 3953 m) in the field and the voucher specimen (Liu2019-53) was deposited in the herbarium of the life college, Lanzhou University. We extracted the total DNAs by adopting the improved CATB method (Doyle and Doyle Citation1987). A short-insert library (270 bp) was constructed and whole-genome sequencing was performed with 150 bp paired-end reads by the Illumina Hiseq 2500 Platform (Illumina, San Diego, CA). We trimmed raw reads and obtained 5 Gb clean reads after quality control by using Fast-Plast v1.2.8 (https://github.com/mrmckain/Fast-Plast). We downloaded the plastome of Rh. palmatum (KR816224) as the reference to assemble the plastome of Rh. likiangense by using NOVOplasty v4.1 (Dierckxsens et al. Citation2016). We further used BWA 48 v.0.7.12 (Li and Durbin Citation2009) and SAMtools v.1.2 (Li et al. Citation2009) to compare the reference sequence with our targeted plastome. We manually adjusted the assembled plastome sequence by Geneious v.R.8.1.4 (Kearse et al. Citation2012). We used ‘plann’ for plastome sequence annotation and correction (Huang and Cronk Citation2015). We illustrated the structural features of the plastome by OGDRAW (Lohse et al. Citation2013) ().
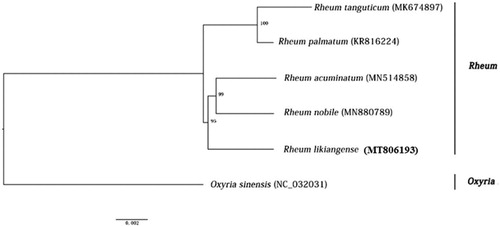
Figure 1. A phylogenetic tree based on the plastome sequences of Rh. likiangense and the closely related species using the Maximum Likelihood method. Bootstrap values are shown above the nodes with 1000 replicates.
The complete plastome of Rh. likiangense is 162,291 bp in length as a quadripartite circle, including a large single copy (LSC) region of 86,853 bp and a small fraction of single copy (SSC) of 11,956 bp, which are separated by two inverted repeat (IR) regions, each 31,741 bp. According to the annotation, this plastome contains 128 genes, including 34 tRNA genes, 8 rRNA genes, and 86 protein-coding genes. There are 90 unique genes and 19 genes are duplicated in the IR regions. The base composition of this genome comprise 31.23% A, 18.93% C, 18.31% G, and 31.53% T, with an overall GC content of 37.24%. The LSC, SSC and IR regions were 35.25%, 32.65% and 40.80%, respectively.
We aligned the total plastome sequences of this and other four rhubarb species (Fan et al. Citation2016; Huo et al. Citation2019; Chen and Li Citation2020; Yang et al. Citation2020) and Oxyria sinensis (Luo et al.Citation2017) using MAFFT v7 (Katoh and Standley Citation2013). We used O. sinensis as the outgoup to perform phylogenetic analyses of five rhubarb species based on the maximum likelihood (ML) approach with the best GTR + F + I model. We carried out bootstrap analyses (1000 times) (Hoang et al. Citation2018) to calculate statistical supports for each clade. The produced ML tree () indicates that Rh. likiangense comprises a well-supported clade with Rh. acuminatum and Rh. nobile, but remains relatively distant from Rh. palmatum and Rh. tanguticum. This interspecific relationship is largely consistent with previous phylogenetic analyses (Wang et al. Citation2005; Sun et al. Citation2012), but with the highly elevated support values.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The chloroplast genome sequence of Rheum likiangense reported here has been submitted to National Center for Biotechnology Information (NCBI) database with an accession number MT806193(https://www.ncbi.nlm.nih.gov/nuccore/MT806193.1/).
Additional information
Funding
References
- Chen Q, Li Y. 2020. The complete chloroplast genome of Rheum nobile. Mitoch DNA Part B. 5(2):1519–1520.
- Dierckxsens N, Mardulyn P, Smits G. 2016. NOVO Plasty: de novo assembly of organelle genomes from whole genome data. Nuc Acid Res. 45:e18.
- Doyle JJ, Doyle CJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fan K, Sun XJ, Huang M, Wang XM. 2016. The complete chloroplast genome sequence of the medicinal plant Rheum palmatum L. Mitoch DNA Part A. 27(4):2935–2936.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522.
- Huang DI, Cronk QC. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Huo X, Wei H, Gao J, Yan Y, Zhang G, Liu M. 2019. The complete chloroplast genome sequence of Rheum tanguticum, an endangered Chinese medicinal plant (Polygonaceae). Mitoch DNA Part B. 4(2):4055–4056.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25(14):1754–1760.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW – a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nuc Acid Res. 41:575–581.
- Luo X, Wang TJ, Hu H, Fan LQ, Wang Q, Hu QJ. 2017. Characterization of the complete chloroplast genome of Oxyria sinensis. Conservation Genet Resour. 9(1):47–50.
- Sun YS, Wang AL, Wan DS, Wang Q, Liu JQ. 2012. Rapid radiation of Rheum (Polygonaceae) and parallel evolution of morphological traits. Mol Phylogenet Evol. 63(1):150–158.
- Wang AL, Yang MH, Liu JQ. 2005. Molecular phylogeny, recent radiation and evolution of gross morphology of the rhubarb genus Rheum (polygonaceae) inferred from chloroplast DNA trnL-F sequences. Ann Bot. 96(3):489–498.
- Xie ZQ. 2000. Studies on certain ecological characteristics of Rheum likiangense in Yushu. Qinghai. Acta Ecol Sin. 20:815–819.
- Yang BB, Li LD, Liu JQ, Zhang LS. 2020. Plastome and phylogenetic relationship of the woody buckwheat Fagopyrum tibeticum in the Qinghai-Tibet Plateau. Pl Diversity. online:
