Abstract
Citrus sunki (Jinkyool) is a medicinal landrace citrus belonging to the Rutaceae family. We determined the complete chloroplast genome (160,699 bp) of C. sunki CRS0085 in Jeju Island, Korea. The genome is composed of four distinct parts; a large single copy of 87,918 bp, a small single copy of 21,355 bp, and a pair of inverted repeat regions of 25,713 bp. A total of 134 genes including 89 protein-coding genes, 37 tRNA genes, and eight rRNA genes were identified. The phylogenetic tree showed that C. sunki CRS0085 has the closest relationship with C. reticulata within genus Citrus.
The subfamily Aurantioideae of the angiosperm family Rutaceae has been classified into two tribes, Clauseneae and Citreae. The genus Citrus and related genera such as Eremocitrus, Fortunella, Microcitrus, and Poncirus belong to the ‘true citrus fruit trees’ of the Citreae tribe (Penjor et al. Citation2013). Modern taxonomic studies have suggested that three citrus types, namely citron (Citrus medica L.), mandarin (C. reticulata Blanco), and pummelo (C. maxima Burm. Merrill) that are fundamental or primary species (Nicolosi Citation2007). According to a recent molecular phylogenetic study, papeda (C. micrantha Wester) that contributed to all limes and lemons is regarded as the fourth basic species (Curk et al. Citation2016).
C. sunki, called Jinkyool in Korean, is a landrace mandarin that grows in Jeju, South Korea (Yi et al. Citation2018), and has been used in traditional Asian medicine to treat various diseases, including indigestion and bronchial asthma (Shin et al. Citation2011). We assembled and annotated the complete chloroplast genome sequence of C. sunki, to provide basic genetic resource for Citrus hybrids with other species in the genus Citrus.
Fresh leaves of C. sunki CRS0085 were obtained from an experimental station of Citrus Research Institute, National Institute of Horticultural & Herbal Science, Republic of Korea (voucher no. IT233734, 33°18′8.21ʺ N/126°36′43.04ʺ E). Total genomic DNA was purified using Biomedic® Plant gDNA Extraction Kit (Biomedic Co., Ltd., Bucheon, Korea) according to the manufacturer's protocol. A genomic library for Nanopore sequencing was constructed, and sequenced using a GridION X5 sequencer (Oxford Nanopore, Oxford, UK) and a TrueSeq DNA library with an average fragment size range of 500 bp was constructed and sequenced using the Illumina HiSeq2500. All sequencing procedures were performed by Phyzen Co. Ltd. (Seongnam, Korea). A total of 8,784,991 Nanopore reads comprising 30.9 Gbp were obtained, and a total of 311,565,668 Illumina reads comprising 47 Gbp were generated. We first assembled Nanopore reads into contigs using SMARTdenovo to obtain putative chloroplast reads. In assembled contigs, we selected putative chloroplast contigs with homology to the CDSs of C. reticulata chloroplast genome (GenBank: NC_034671.1) using TBLASTN, and mapped Illumina reads and Nanopore reads to the chosen contigs using Bowtie2 (Langmead and Salzberg Citation2012) and Minimap2 (Li Citation2018), respectively. The resulting aligned reads were co-assembled using Unicycler (ver. 0.4.6) (Wick et al. Citation2017), and one circular contig was produced. The genome was annotated by GeSeq (Tillich et al. Citation2017), followed by manual confirmation using BLAST searches.
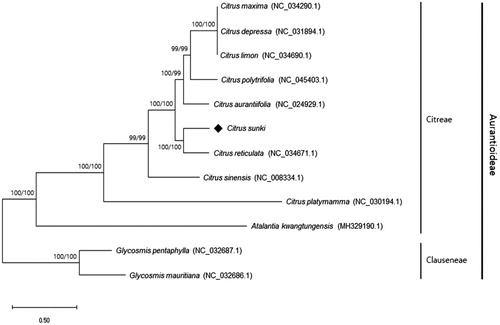
The complete chloroplast genome of C. sunki CRS0085 (GenBank accession no. MT767776) was 160,699 bp in length, and separated into four distinct regions: a large single copy (LSC) (87,918 bp), a small single copy (SSC) (21,355 bp), and a pair of inverted repeats (IRa and IRb) (each 25,713 bp). In the chloroplast genome, a total of 134 genes including 89 protein-coding genes, 37 tRNA genes, and eight rRNA genes were identified. The aligned complete chloroplast genome sequences of C. sunki CRS0085 and those of 11 Aurantioideae species were used for phylogenetic analysis by MEGA-X (Kumar et al. Citation2018). The phylogenetic tree was constructed using the maximum likelihood method with 1000 bootstrap replications, in which C. sunki CRS0085 was clustered with C. reticulata and mostly close to another seven Citrus species (). Phylogenetic analysis using matK gene sequences also showed that C. sunki belonged to the mandarin cluster (Penjor et al. Citation2013). The complete chloroplast genome sequence of C. sunki can be utilized for DNA barcoding and evolutionary studies of the genus Citrus.
Disclosure statement
Authors declare that none of the authors have any competing interests in the manuscript.
Data availability statement
The data that support the findings of this study were deposited to the NCBI GenBank database (accession no. MT767776) (https://www.ncbi.nlm.nih.gov/nuccore/MT767776).
Additional information
Funding
References
- Curk F, Ollitrault F, Garcia-Lor A, Luro F, Navarro L, Ollitrault P. 2016. Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann Bot. 117(4):565–583.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 34(18):3094–3100.
- Nicolosi E. 2007. Origin and taxonomy. In: Khan IA, editor. Citrus genetics, breeding and biotechnology. Oxfordshire (UK): CAB International; p. 19–43.
- Penjor T, Yamamoto M, Uehara M, Ide M, Matsumoto N, Matsumoto R, Nagano Y. 2013. Phylogenetic relationships of Citrus and its relatives based on matK gene sequences. PLoS One. 8(4):e62574.
- Shin HS, Kang SI, Ko HC, Kim HM, Hong YS, Yoon SA, Kim SJ. 2011. Anti-inflammatory effect of the immature peel extract of Jinkyool (Citrus sunki Hort. ex Tanaka). Food Sci Biotechnol. 20(5):1235–1241.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 13(6):e1005595.
- Yi KU, Kim HB, Song KJ. 2018. Karyotype diversity of Korean landrace mandarins by CMA banding pattern and rDNA loci. Sci Hortic. 228:26–32.