Abstract
The Japanese Glandirana rugosa phylogenetically consists of four groups. However, the taxonomic identity of these groups still remains unclear. We determined the complete mitogenome sequences of the four groups of G. rugosa. The mitogenomes were 17,394–17,781 bp in length. The phylogenetic analysis clearly showed that the genus Glandirana is monophyletic and that the four groups of G. rugosa are separated into two clusters: one cluster represents G. rugosa, the other cluster may represent a different species.
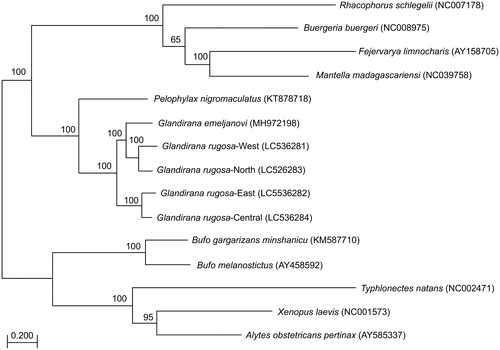
The Japanese Glandirana rugosa was one of the five frog species collected by Siebold during his stay in Nagasaki (Temminck and Schlegel Citation1838). Originally, this frog was described as Rana rugosa Schlegel in Temminck and Schlegel (Citation1838). Later, the species was transferred into the genus Glandirana (Frost Citation2013). The Glandirana rugosa is now separated into four groups, designated as the West, East, North, and Central groups, based on the mitochondrial 12S rRNA gene (Oike et al. Citation2017). This suggests that the Japanese G. rugosa probably consists of cryptic species. We collected individuals of G. rugosa in the Cities of Higashi-Hiroshima (West; 34°29′03.6″N 132°42′13.4″E), Nagaoka (North; 37°21′32.7″N 138°55′26.1″E), Shizuoka (Central; 35°06′28.1″N 138°21′51.2″E), and Kamogawa (East; 35°08′22.8″N 140°12′32.2″E). We stored the tissues in Dr. Nakamura lab, Ehime University (sample database number: NSE_A00101-4). DNA was extracted (mtDNA extractor CT kit, Wako, Osaka, Japan) and cleaved with PstI and EcoRI which yielded suitable DNA size fragments for cloning. Cloned DNA fragments were Sanger sequenced (following Sumida et al. Citation2001) and the mitogenome of Pelophylax nigromaculatus (GenBank ACCN AB043889) was used as reference for assembling. The complete mitogenomes of the four groups of G. rugosa (GenBank ACCN LC536281-4) were 17,394–17,781 bp in length, consisting of 13 protein-coding genes, 12S and 16S rRNAs, 21 tRNAs, and one pseudo-tRNA and one non-coding D-loop. The RAxML () and MrBayes phylogenetic trees revealed the monophyly of the genus Glandirana in which G. rugosa-West and -North, and G. emeljanovi were separated from G. rugosa-East and -Central (); however, the latter tree is not shown because of a figure limit. The Korean G. emeljanovi is probably a different species from the Japanese G. rugosa, based on the relatively low genetic identity between G. emeljanovi and G. rugosa (probably West group) mitogenomes (84.0%) given the range of intraspecific genetic identity in Ranidae frogs is 94.0–100.0% (Eo et al. Citation2019). In this study, we found that genetic identity values of mitogenomes of G. rugosa-North, -East, -Central, and G. emeljanovi were 88.6, 81.8, 82.4, and 84.8%, respectively, when the G. rugosa-West was set as 100%. We also show homology of nucleotide sequences of 13 protein-coding genes among the four groups in Table S1. From these results, we concluded that G. rugosa-West and -North are a different species from G. rugosa-East and -Central, and from the Korean G. emeljanovi. Thus, the Japanese G. rugosa is separated into at least two independent species. The West group likely represents G. rugosa s.str., because it contains individuals from the supposed type locality, Nagasaki (Oike et al. Citation2017). We argue that the East and Central groups should have a new species name.
tmdn_a_1833772_sm6280.xlsx
Download MS Excel (9.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data are available at the website (https://www.ncbi.nlm.nih.gov/genbank/) under ACCN LC536281-4.
Additional information
Funding
References
- Eo SH, Lee B-J, Park C-D, Jung J-H, Hong N, Lee W-S. 2019. Taxonomic identity of the Glandirana emeljanovi (Anura, Ranidae) in Korea revealed by the complete mitochondrial genome sequence analysis. Mitochondrial DNA B. 4(1):961–962.
- Frost DR. 2013. Amphibian species of the world: an online reference. Version 5.6. http://research.amnh.org/herpetology/amphibia/index.html.
- Oike A, Watanabe K, Min M-S, Tojo K, Kumagai M, Kimoto Y, Yamashiro T, Matsuo T, Kodama M, Nakamura Y, et al. 2017. Origin of sex chromosomes in six groups of Rana rugosa frogs inferred from a sex-linked DNA marker. J Exp Zool. 327(7):444–452.
- Sumida M, Kanamori Y, Kaneda H, Kato Y, Nishioka M, Hasegawa M, Yonekawa H. 2001. Complete nucleotide sequence and gene rearrangement of the mitochondrial genome of the Japanese pond frog Rana nigromaculata. Genes Genet Syst. 76(5):311–325.
- Temminck CJ, Schlegel H. 1838. Reptilia. In: de Siebold PF, editor. Fauna Japonica. Amsterdam: Lugduni Batavorum; p. 110–111.