ABSTACT
Phalaenopsis mannii, one of the native Phalaenopsis in China, is an important parent for breeding new varieties. However, its position has been unclear in Phalaenopsis. The obtained high-quality P. mannii chloroplast genome will provide useful information for phylogenetic and future breeding of Phalaenopsis. Herein, we reported a complete chloroplast genome of P. mannii from Yunnan, China. The sequencing data obtained from BGISEQ-500 platform were assembled. This sequence had a circular molecular length of 148,596 bp and contained a total of 127 genes with an average GC content of 36.7%. Phylogenetic analysis indicated that Phalaenopsis was monophyletic with strong support, in which the P.mannii was the sister-group of Phalaenopsis aphrodite subsp. formosas, Phalaenopsis ‘TinyStar’ and Phalaenopsis equestris.
Phalaenopsis, as a member of orchid family, is famous for its high ornamental characteristics and unique taxonomic status. It is also highly regarded by many scholars. However, the taxonomic position of some species in Phalaenopsis are still unclear. Phalaenopsis mannii, which is used as an important parent in cross breeding, is an epiphytic orchid native to southern Yunnan province, China (Chen and Ji Citation2000). Previously, largely according to flower and other morphological characteristics, P. mannii belongs to the sect. Polychilos. But phylogenetic trees constructed based on nuclear or chloroplast genes manifested that P. mannii was embedded in sect. Amboinenses, and sometimes it was the sister relationship to sect. Amboinenses and sect. Zebrinae as a single branch (Ito et al. Citation2005; Tsai et al. Citation2010). Thus, it is interesting to assemble and characterize the P. mannii chloroplast genome for providing a better studying phylogenetic and future breeding of Phalaenopsis.
Fresh samples of P. mannii was collected from Yunnan province, China (23°12′N, 104°70′E), and the living plants deposited in Fujian Agriculture and Forestry University (26°04′51.3″N, 119°14′19.9″E, Voucher specimen FAFU: HDL-YN2019-12A). Total genomic DNA from fresh P. mannii leaves was extracted with the DP305-Plant Tissues Genome DNA Extraction Kit (TianGen, Beijing, China) according to the kit instructions. The DNA sheared to approximately 300-500 bp. Then the libraries were constructed through end-repair, A-tailing, adapter ligation. Approximately 20 Gb clean reads were obtained through the BGISEQ-500 platform sequenced. After adapters and low-quality reads were removed by fastp software (Mak et al. Citation2017; Chen et al. Citation2018). Draft chloroplast genome was obtained using SPAdes v 3.13.1 software and manually corrected using Bandage v 0.8.1 software (Zhou et al. Citation2019). The assembled chloroplast genome was annotated by the online tool Geseq (Tillich et al. Citation2017) and Geneious v 2020.2.1 software (reference: P. ‘Tiny Star’, KJ944326) and then checked manually. In the end, we established the complete chloroplast genome sequence of P. mannii (GenBank accession MT822270) with a circular molecule of 148,596 bp in length and overall GC content of 36.7%. This chloroplast genome included an LSC region of 85,300 bp, an SSC region of 11,640 bp, and a pair of inverted repeats regions of 25,828 bp. We annotated 127 genes for the chloroplast genome, including 76 protein-coding genes (PCG), 37 transfer RNAs (tRNAs), eight ribosomal RNA (rRNA) genes, and six pseudogenes.
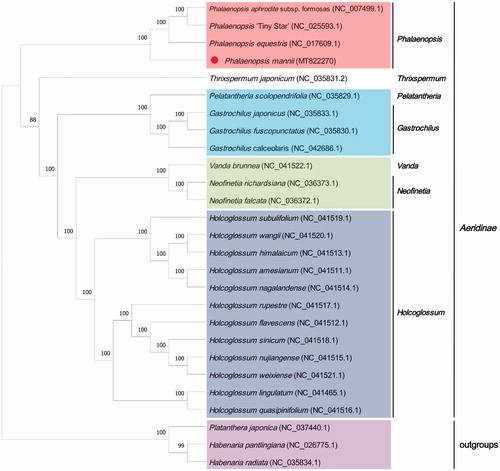
The phylogenetic tree was constructed to demonstrate the relationships between Phalaenopsis and other genera. The P.mannii chloroplast genome was aligned with other 24 Aeridinae chloroplast genome sequences and three Orchidinae species as the outgroup by Mafft v 7.455, and the construct the maximum likelihood tree by RAxML v 8.2.12 (Katoh and Standley Citation2013; Stamatakis Citation2014). All the sequences were downloaded from NCBI GenBank. The maximum likelihood tree indicated that Phalaenopsis formed a monophyletic clade, in which the P.mannii was the sister-group of (Phalaenopsis equestris, (Phalaenopsis aphrodite subsp. formosas, Phalaenopsis ‘TinyStar’)) ().
Acknowledgments
The authors are grateful to Jieyu Wang, Yang Hao and Xiongde Tu for providing the information of the plant material.
Disclosure statement
There no conflicts of interest for all the authors. The authors alone are responsible for the content and writing of the article.
Data availability statement
The raw data has been stored in omics database of Genome Sequence Archive. GSA accession number is CRA003144. All the information can be found on the website (https://bigd.big.ac.cn/gsa/browse/CRA003144/CRX150834).
Additional information
Funding
References
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Chen XQ, Ji ZH. 2000. Chinese Orchids. Beijing: China Forest Industry press.
- Ito M, Kita K, Handa T, Hidayat T, Yukawa T. 2005. Molecular phylogenetics of “Phalaenopsis” (Orchidaceae) and allied genera: re-evaluation of generic concepts. Acta Phytotaxonomica et Geobotanica. 56:141–162.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Mak SST, Gopalakrishnan S, Carøe C, Geng C, Liu S, Sinding M-HS, Kuderna LFK, Zhang W, Fu S, Vieira FG, et al. 2017. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience. 6(8):1–13.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Tsai CC, Chiang YC, Huang SC, Chen CH, Chou CH. 2010. Molecular phylogeny of Phalaenopsis Blume (Orchidaceae) on the basis of plastid and nuclear DNA. Plant Syst Evol. 288(1–2):77–98.
- Zhou Y, Chen B, Zheng Y, Wei Z, Zhao K. 2019. Basic chloroplast characterizations of Prunus campanulata x kanzakura ‘praecox’, a warm-adapted cherry cultivar in South China (Rosaceae). Mitochondrial DNA B. 4(2):3945–3947.