Abstract
Euphorbia maculata is an important medicinal plants of the family Euphorbiaceae. The complete chloroplast genome reported here is 162,685 bp in length, including two inverted repeats (IRs) of 26,822 bp, which are separated by a large single-copy (LSC) and a small single-copy (SSC) of 90,514 bp and 18,527 bp, respectively. The whole chloroplast genome of E. maculata contains 111 genes, including 77 protein-coding genes, 4 transfer RNA, and 30 ribosome RNA. Phylogenetic analysis indicated that E. maculata is closely related to E. milii and E. tirucalli.
Euphorbia maculata L. (Spotted spurge) is a fast-growing annual weed native to eastern North America and spread to Asia, Africa and Europe (Wu et al. Citation1994). It functions as a pioneer species with a prostrate growth habit in ecological succession. The milky sap of the plant is an irritant for many people. Like many members of the family Euphorbiaceae, it has been widely used as a folk medicine, which can produce anti-inflammatory and cancer chemopreventive agents of triterpenoids (Yi et al. Citation2018). It is necessary to develop genomic resources for E. maculata to provide intragenic information for its utilization and chloroplast genomes are valuable sources (Dong et al. Citation2020; Sun et al. Citation2020).
The fresh leaves of E. maculata were collected from Luxi county, Hunan province, China (28°12’59”N, 110°13’11”E). Voucher specimens were deposited in Institute of Chinese Materia Medica (Specimen accession number: 430723LY0485), China Academy of Chinese Medical Sciences. Total genomic DNA was extracted with the modified cetyltrimethyl ammonium bromide (CTAB) method (Li et al. Citation2013). Paired-end libraries were prepared with the NEBNext Ultra DNA Library Prep Kit. The genome was sequenced using the HiSeq X Ten platform (Illumina, Santiago, CA, USA). All good quality paired reads were assembled using the Spades program to contigs (Bankevich et al. Citation2012). Chloroplast genome sequence contigs were selected by the program BLAST (Altschul et al. Citation1990) using E. milii (Genbank accession number: MN713924) as a reference and the selected contigs were assembled using Sequencher 4.10 (Gene Codes Corporation, Ann Arbor, MI USA, http://www.genecodes.com). Chloroplast genome annotation was performed with Plann (Huang and Cronk Citation2015) using the E. milii as reference sequence. The annotated sequence was submitted to the GenBank under the accession number MT830858.
The complete chloroplast genome reported here is 1,62,685 bp in length, including two inverted repeats (IRs) of 26,822 bp, which are separated by a large single-copy (LSC) and a small single-copy (SSC) of 90,514 bp and 18,527 bp, respectively. The overall GC-content of the chloroplast genome was 35.4%. The chloroplast DNA of E. maculata comprised 111 distinct genes, including 77 protein-coding genes, 4 transfer RNA, and 30 ribosome RNA, but didn’t contain cemA and rpl22 these two protein-coding genes. In these genes, 17 harbored a single intron, while two (ycf3and clpP) contained double introns.
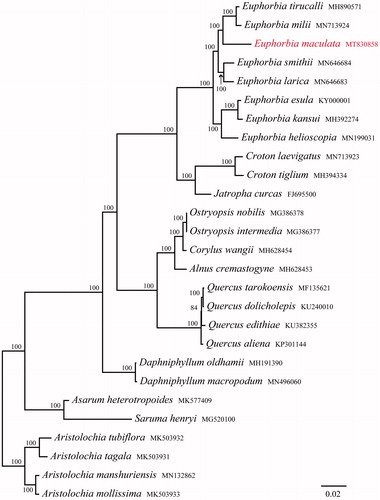
In order to confirm the phylogenetic relationships of E. maculata within the genus Euphorbia and other related groups, total 26 complete cp genomes were obtained from Genbank and the genus Aristolochia was taken as an outgroup. All chloroplast genome sequences were aligned using MAFFT (Katoh et al. Citation2019) and ambiguous alignment regions were trimmed by Gblocks (Castresana Citation2000). Phylogenetic analysis was conducted based on maximum-likelihood (ML) analyses using RAxML (Stamatakis Citation2014), under the GTR + G model with 1000 rapid bootstrap replicates. The phylogenetic tree showed that all species of Euphorbia form a monophyletic group with 100% support, and E. maculata is closely related to E. milii and E. tirucalli (). The chloroplast genome of E. maculata provided a lot of genetic information for species conservation and identification of genus Euphorbia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI https://www.ncbi.nlm.nih.gov/, reference number MT830858, raw data BioProject ID: PRJNA662166.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Castresana J. 2000. GBLOCLKS: selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Version 0.91b. Mol Biol Evol. 17:540–552.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Dong W, Xu C, Wen J, Zhou S. 2020. Evolutionary directions of single nucleotide substitutions and structural mutations in the chloroplast genomes of the family Calycanthaceae. BMC Evol Biol. 20(1):96.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Li JL, Wang S, Jing Y, Wang L, Zhou SL. 2013. A modified CTAB protocol for plant DNA extraction. Chin Bull Bot. 48:72–78.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Sun J, Wang Y, Liu Y, Xu C, Yuan Q, Guo L, Huang L. 2020. Evolutionary and phylogenetic aspects of the chloroplast genome of Chaenomeles species. Sci Rep. 10(1):11466.
- Wu ZY, Raven PH, Hong DY. 1994. Flora of China. 44.
- Yi S, Liang-Liang G, Meng-Yue T, Bao-Min F, Yue-Hu P, Ken Y. 2018. Triterpenoids from euphorbia maculata and their anti-inflammatory effects. Molecules. 23:2112.