Abstract
The genus of Coelogyne Lindl. comprised about 200 species while its generic relationship has been uncertain. The whole chloroplast genome of C. barbata was reported in order to provide new data on the molecular phylogeny of Coelogyne. The cp genome of C. barbata was 1,600,93 bp in total length, including a pair of inverted repeat regions (IR, 26,710 bp), one large single-copy region (LSC, 87,868 bp), and one small single-copy region (SSC, 188,05 bp). The complete chloroplast DNA encoded 132 genes, containing 86 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Phylogenetic analysis showed that C. barbata was related to Pholidota imbricata.
The genus of Coelogyne Lindl. (tribe Arethuseae, subfam. Epidendroideae) comprised about 200 species in which generic relationship had been uncertain (Chase et al. Citation2015). C. barbata Lindl. ex Griff. was recognized by possessing sparse pseudobulbs with two leaves, hysteranthous inflorescence with white flowers, and margin long fimbriate on the lip. It grew on trees in forests or on cliffs and was distributed in SW China, Bhutan, NE India, and Nepal (Chen and Clayton Citation2009). Since the plastid genome would provide molecular evidence for plant systematic (Jansen et al. Citation2007; Nock et al. Citation2011; Pan et al. Citation2019), the complete chloroplast genome of C. barbata was reported.
Leaf samples of C. barbata were obtained from Fumin County, Yunnan Province, China (25°20′19″N, 102°27′26″E). The voucher specimen was deposited in the Herbarium of Southwest Forestry University (HSFU, Lilu-20180004). High quality chloroplast genomic DNA of C. barbata was extracted from the fresh leaf by using the modified CTAB procedure (Doyle and Doyle Citation1987) and sequenced on the Illumina Hiseq 2500 platform (Illumina, San Diego, CA) in Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). Genome sequences were assembled and screened using GetOrganelle pipe-line (Jin et al. Citation2018). Then, the new sequence was annotated with Geneious Prime version 2020.1.2 (Kearse et al. Citation2012). Finally, the chloroplast genome sequence was submitted to GenBank and obtained an access number MT627604.
The complete chloroplast genome of C. barbata was 160,093 bp in length and contained two inverted repeats (IR, 26,710 bp) regions, a large single-copy region (LSC, 87,868 bp), and a small single-copy region (SSC, 18,805 bp). This gene sequence encoded 132 genes, including 86 protein-coding genes, 38 tRNA genes, and eight rRNA genes. Among them, LSC, SSC, and IR regions accounted for 35.2%, 30.5%, and 43.3% of the GC-content. In addition, the overall GC-content was 37.3%. The gene order and organization of C. barbata were consistent with those of Dendrobium species in subfamily Epidendroideae (Wang et al. Citation2020).
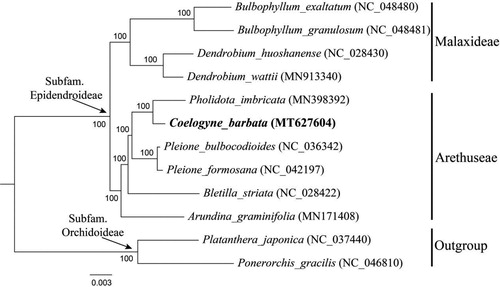
To confirm the phylogenetic position of C. barbata, a maximum likelihood (ML) tree was conducted using RAxML v8.1.11 (Stamatakis Citation2014) as implemented on the Cyberinfrastructure for Phylogenetic Research (CIPRES) Science Gateway (http://www.phylo.org/, Miller et al. Citation2010), employing the GTRtG model with 1000 bootstrap iterations (-#/-N). Other parameters used the default settings. Phylogenetic analysis based on 79 protein-coding genes of 10 representative plastomes within the subfamily Epidendroideae as ingroup. According to the updated molecular phylogeny of Orchidaceae (Chase et al. Citation2015; Li et al. Citation2016), 10 species from two tribes (Malaxideae Lindl. and Arethuseae Lindl.) in subfamily Epidendroideae were selected as ingroup, while two species from subfamily Orchidoideae as outgroup. It turned out that C. barbata was closely related to Pholidota imbricata Hook. with 100% support ().
Acknowledgements
First of all, we were grateful to Dr. Fei Zhao in Kunming Institute of Botany Chinese Academy of Sciences for his help in this article. Second, we were grateful to Mr. Zhi-feng Xu and Mrs. Xiao-yun Wang in the Wild Orchid Conservation Center of Yunnan Fengchunfang Biotechnology Company for providing the materials.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI at https://www.ncbi.nlm.nih.gov, reference number MT627604.
Additional information
Funding
References
- Chase MW, Cameron KM, Freudenstein JV, Pridgeon AM, Salazar G, Berg C, Schuiteman A. 2015. An updated classification of Orchidaceae. Bot J Linn Soc. 177(2):151–174.
- Chen SC, Clayton D. 2009. Coelogyne Lindley. In: Wu ZY, Raven PH, Hong D, editors. Flora of China. Vol. 25. Beijing (China); St. Louis (MO): Science Press; Missouri Botanical Garden Press; p. 315–325.
- Doyle JJ, Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 104(49):19369–19374.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. BioRxiv. 256479.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li MH, Zhang GQ, Lan SR, Liu ZJ. 2016. A molecular phylogeny of Chinese orchids. J Syst Evol. 54(4):349–362.
- Miller MA, Wayne P, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Paper presented at: 2010 Gateway Computing Environments Workshop (GCE). IEEE Computer Society; p. 1–8.
- Nock CJ, Waters DL, Edwards MA, Bowen SG, Rice N, Cordeiro GM, Henry RJ. 2011. Chloroplast genome sequences from total DNA for plant identification. Plant Biotechnol J. 9(3):328–333.
- Pan YY, Li TZ, Chen JB, Huang J, Rao WH. 2019. Complete chloroplast genome of Dendrobium thyrsiflorum (Orchidaceae). Mitochondrial DNA B. 4(2):3192–3193.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England). 30(9):1312–1313.
- Wang YP, Ai J, Luo Y, Li QQ, Li L. 2020. The complete chloroplast genome of Dendrobium wattii (Orchidaceae). Mitochondrial DNA B. 5(2):1325–1326.