Abstract
We determined the complete mitochondrial genome of Lactoria cornuta, which is 16,495 bp in length with an A + T content of 57.37%, and contains 13 protein-coding genes, 2 rRNAs, 22 tRNAs and a complete control region. The total base composition of the mitogenome is 28.2% T, 26.7% C, 29.2% A and 15.9% G. Of the 13 protein-coding genes, 12 genes start with an ATG codon, except for COX1 with GTG. Ten genes use TAA or AGA as the termination codon, whereas three (COX2, ND4, and Cyt b) have incomplete stop codon T. This study would provide useful genetic information for phylogenetic and species idenfication of the family Ostraciidae.
Lactoria cornuta is a small box-like fish belongs to the family Ostraciidae, which are widely distributed in coral reefs from the Indian Ocean to the Pacific Ocean. When threatened or harassed, they can exude ostracitoxin. In spite of their peculiar morphology, the family Ostraciidae have received relatively scant attention from fish systematists, and the relationships within genera appeared to be uncertain (Santini et al. Citation2013). Until now, only rare studies have reported the complete mitochondrial genome of the family Ostraciidae, such as Lactoria diaphana (NC_011330; Yamanoue et al. Citation2008), O. immaculatus (NC_009865; Yamanoue et al. Citation2007) and Ostracion rhinorhynchos (KU308378; Huang et al. Citation2016). Hence, we sequenced its mitochondrial genome of L. cornuta and analyzed its phylogenetic position.
The specimen of L. cornuta was collected from Guangdong Lanhai Marine Technology Co., Ltd, Guangzhou City, Guangdong Province, China (23° 12′51″N, 113° 28′06″E). Genomic DNA was extracted from muscle using Tissue DNA Kit (OMEGA E.Z.N.A). The samples were stored in −80 °C in National Freshwater Aquatic Germplasm Resource Center in Guangzhou city, Guangdong Province in China. Number is OS-LC-1. The mitogenome was sequenced by the next-generation sequencing using an Illumina HiSeq 2000 system and assembled using SPAdes v.3.5.0 software (http://cab.spbu.ru/software/spades/) (Lapidus et al. Citation2014).
The complete mitogenome of L. cornuta was 16,495 bp in length, containing 22 transfer RNA genes (tRNAs), 13 protein-coding genes (PCGs), 2 ribosome RNA genes (rRNAs), and a control region (CR), which was similar to those of the family Ostraciidae (Yamanoue et al. Citation2007; Huang et al. Citation2016). The total nucleotide composition is a 28.2% T, 26.7% C, 29.2% A and 15.9% G, respectively. Of the 13 protein-coding genes, 12 genes start with an ATG codon, except for COX1 with GTG. Ten use TAA or TAG as the termination codon, whereas three (COX2, ND4, and Cyt b) have incomplete stop codon T.
Among the 13 PCGs ranging in size from 168 bp (ATPase8) to 1839 bp (ND5), 12 PCGs are coded on the H-strand, while ND6 is coded on the L-strand. The 12S and 16S rDNA are 949 bp and 1687 bp in length, respectively. The length variations of 22 tRNAs range from 67 bp (tRNACys) to 75 bp (tRNALys). There are two non-coding regions, the L-strand replication origin region (38 bp) locating between tRNAAsn and tRNACys, and the control region (833 bp) locating within the tRNAPro and tRNAPhe.
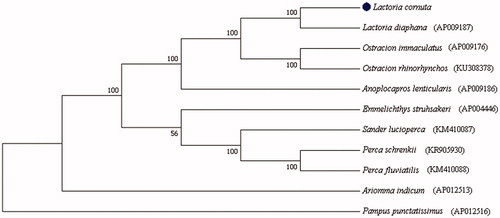
To determine taxonomic status of L. cornuta, we reconstructed the phylogeny of the family Ostraciidae based on the nucleotide sequence of 13 protein-coding genes in the mitochondrial genome. Phylogenetic analyses highly supported the close relationship of L. cornuta and L. diaphana. The genus Lactoria and the genus Ostracion were clustered (), consistent with previous sutdy (Yamanoue et al. Citation2008). The complete mitochondrial genome sequence of L. cornuta would provide useful genetic markers for population genetics and species identification of fish.
Disclosure statement
The authors report no conflicts of interests. The authors alone are responsible for doing the research and writing the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, accession number MT712150.
Additional information
Funding
References
- Huang Y, Zhao X, Ruan Z, Wang M, Xu J, Shi Q. 2016. The complete mitochondrial genome of horn-nosed boxfish (Ostracion rhinorhynchos). Mitochondrial DNA Part B. 1(1):100–102.
- Lapidus A, Antipov D, Bankevich A, Gurevich A, Korobeynikov A, Nurk S, Prjibelski A, Safonova Y, Vasilinetc I, Pevzner PA. 2014. New Frontiers of Genome Assembly with SPAdes 3.0. (poster). http://cab.spbu.ru/software/spades/
- Santini F, Sorenson L, Marcroft T, Dornburg A, Alfaro ME. 2013. A multilocus molecular phylogeny of boxfishes (Aracanidae, Ostraciidae; Tetraodontiformes). Mol Phylogenet E. 66(1):153–160.
- Yamanoue Y, Miya M, Matsuura K, Katoh M, Sakai H, Nishida M. 2008. A new perspective on phylogeny and evolution of tetraodontiform fishes (Pisces: Acanthopterygii) based on whole mitochondrial genome sequences: basal ecological diversification? BMC Evol Biol. 8(1):212–212.
- Yamanoue Y, Miya M, Matsuura K, Yagishita N, Mabuchi K, Sakai H, Katoh M, Nishida M. 2007. Phylogenetic position of tetraodontiform fishes within the higher teleosts: Bayesian inferences based on 44 whole mitochondrial genome sequences. Mol Phylogenet E. 45(1):89–101.