Abstract
The complete mitogenome of Lasioglossum affine (Hymenoptera: Halictidae) was sequenced and analyzed. The whole mitogenome is 17,352 bp (AT%=84.1%) and encodes 37 typical eukaryotic mitochondrial genes, including 13 protein-coding genes (PCGs), 22 tRNAs, two rRNAs, and an AT-rich region. Further analysis found three gene rearrangements, where trn I-Q-M → trn M-I-Q, trn W-C-Y → trn C-W-Y, and trn K-D → trn D-K were shuffled. The phylogenetic relationships of 19 species of Hymenoptera were established using maximum-likelihood method based on 13 concatenated PCGs. The result showed that Lasioglossum affine is a sister of Lasioglossum sp. SJW-2017.
Lasioglossum affine, also known as Evylaeus affine (Smith 1853), belongs to genus Lasioglossum of the family Halictidae (Sakagami et al. Citation1982). It is a native species in Heilongjiang, Taiwan, Hunan of China, and also distributes in Japan, South Korea, North Korea, and Russia (Sakagami et al. Citation1982; Packer Citation1991; Murao et al. Citation2014). Up to now, little systematic research on the genus Lasioglossum in China has been conducted. Therefore, this gap needs to be addressed (Zhang et al. Citation2012). To explore the genus phylogeny, the first complete mitogenome of L. affine was sequenced and analyzed. The specimen was collected for the first time in Longping village, Cili county, Hunan of China (N 29.450217, E 110.096235). The samples used in this study are stored in the Key Laboratory of Vector Insects in Chongqing Normal University (No. 2018-HN-807). GenBank accession number of this mitogenome is MT780494.
A pair-end (PE) library was built and mitochondrial DNA sequencing was implemented on Illumina Nova seq 6000 platform (Illumina, San Diego, CA). Subsequently, de novo assembly and annotation of complete mitochondrial sequence were carried out using SPAdes v3.9.0 (Bankevich et al. Citation2012) and MITOS (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013), respectively. Finally, the complete mitochondrial genome of L. affine was assembled as a circular DNA molecule of 17,352 bp, encoding 13 protein-coding genes (PCGs), 22 tRNAs, two rRNAs, and an AT-rich region (also called control region, CR). For whole mitogenome, AT content is 84.1%. Compared with the ancestral sequence of Chalcidoidea (Hymenoptera) (Zhu et al. Citation2018), three gene rearrangements were found: trn I-Q-M → trn M-I-Q, trn W-C-Y → trn C-W-Y, and trn K-D → trn D-K. Additionally, nine overlapping regions between gene sequences were scattered throughout the whole mitochondrial genome.
Four types of start codons of 13 PCGs are used: ATA (cox2, nad5, cytb, and nad1), ATT (cox1, cox3, nad3, nad4l, and nad6), ATC (nad2 and atp8), and ATG (atp6 and nad4). The stop codon of all PCGs is TAA. Except for trnS2, all tRNA genes have a typical clover structure and the dihydrouridine (DHU) arm forms a simple loop, which is also conserved in other Hymenoptera species (Wei et al. Citation2010; Huang et al. Citation2016; Lu and Huang Citation2020). The 16S rRNA (rrnL) (1394 bp) and 12S rRNA (rrnS) (842 bp) genes are located on the light strand. The AT-rich region is 2311 bp.
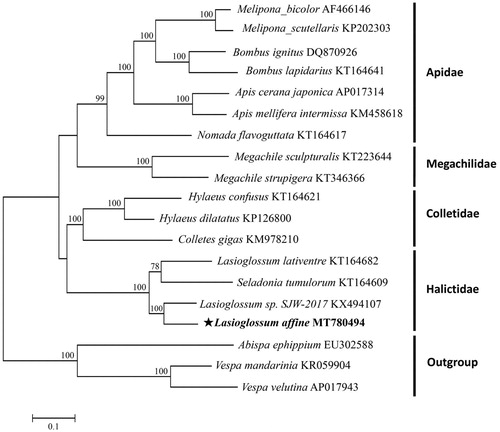
To explore the phylogeny of L. affine in order Hymenoptera, 18 complete mitogenome sequences were downloaded from GenBank, including three outgroups from family Vespidae (Vespa velutina, Vespa mandarinia, and Abispa ephippium). The phylogenetic tree was constructed using 13 concatenated PCGs based on the optimized evolutionary model of GTR + G+I, and the maximum-likelihood (ML) tree was built using MEGA7 with 1000 bootstrap replicates (Kumar et al. Citation2016).
As shown in , the phylogenetic relationships of all tested species were consistent with previous studies (He et al. Citation2018; Lu and Huang Citation2020). L. affine was a sister of Lasioglossum sp. SJW-2017. Taken together, the newly sequenced mitochondrial genome of L. affine can provide DNA barcodes for phylogenetic and evolutionary analysis among other insect species.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitochondrial genome sequence used in this study can be obtained on NCBI through GenBank No. (NCBI https://www.ncbi.nlm.nih.gov/), and raw data were also openly available at SRA (https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP278000).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- He B, Su TJ, Wu YP, Xu JS, Huang DY. 2018. Phylogenetic analysis of the mitochondrial genomes in bees (Hymenoptera: Apoidea: Anthophila). PLOS One. 13(8):e0202187.
- Huang DY, Su TJ, He B, Gu P, Liang AP, Zhu CD. 2016. Sequencing and characterization of the Megachile strupigera (Hymenoptera: Megachilidae) mitogenome. Mitochondrial DNA Part B. 1(1):282–284.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Lu HH, Huang DY. 2020. The complete mitogenome of Habropoda rodoszkowskii (Hymenoptera: Apidae) and phylogenetic analysis. Mitochondrial DNA Part B. 5(3):2350–2351.
- Murao R, Heung-Sik L, Tadauchi O. 2014. Lasioglossum (Acanthalictus) dybowskii (Hymenoptera, Halictidae) newly recorded from South Korea, with a checklist of the genus Lasioglossum in Korean Peninsula. J Hymenoptera Res. 38:141–153.
- Packer L. 1991. The evolution of social behavior and nest architecture in sweat bees of the subgenus Evylaeus (Hymenoptera: Halictidae): a phylogenetic approach. Behav Ecol Sociobiol. 29(3):153–160.
- Sakagami S, Hirashima Y, Maeta Y, Matsumura T. 1982. Bionomic notes on the social halictine bee Lasioglossum affine (Hymenoptera, Halictidae). Esakia. 19:161–176.
- Wei SJ, Tang P, Zheng LH, Shi M, Chen XX. 2010. The complete mitochondrial genome of Evania appendigaster (Hymenoptera: Evaniidae) has low A + T content and a long intergenic spacer between atp8 and atp6. Mol Biol Rep. 37(4):1931–1942.
- Zhang R, Niu ZQ, Li Q, Zhu CD. 2012. Two new species of subgenus Lasioglossum (Hymenoptera: Halictidae) from China. Acta Zootaxon Sin. 37(2):370–373.
- Zhu JC, Tang P, Zheng BY, Wu Q, Wei SJ, Chen XX. 2018. The first two mitochondrial genomes of the family Aphelinidae with novel gene orders and phylogenetic implications. Int J Biol Macromol. 118(Pt A):386–396.