Abstract
The two complete mitochondrial genomes were sequenced from the freshwater monogonont rotifer Brachionus angularis. The mitochondrial genome sequences were 10,764 bp (mitochondrial DNA I) and 12,238 bp (mitochondrial DNA II) in size, respectively. The gene structure and its orientation of 12 protein-coding genes (PCGs) of complete mitochondrial genomes of B. angularis was identical to those shown in other marine rotifers and the freshwater rotifer Brachionus rubens, but was different from the freshwater rotifer Brachionus calyciflorus. Of 12 PCGs, one gene (ND5) had incomplete stop codon. Furthermore, the start codon for CO1, ND4L, ND5, and CO2 was GTG, while the start codon for ND3 and other PCGs was ATA and ATG, respectively. The base composition of 12 PCGs in B. angularis mitogenome showed 20.4% for A, 47.3% for T, 17.5% for C, and 14.8% for G, respectively. The mitochondrial genome A + T base composition (67.7%) of 12 PCGs was higher than G + C (32.3%), while the complete mitochondrial genome A + T base composition (66.3%) was higher than G + C (33.7%).
The freshwater rotifer Brachionus angularis consists of at least four subspecies (Brachionus angularis angularis, Brachionus angularis bidens, Brachionus angularis caudatus, and Brachionus angularis dolabratus) (Hu et al. Citation2003; Segers Citation2007; Hu and Xi Citation2008). However, to date, there is no report on complete mitochondrial genome of B. angularis, while several complete mitochondrial genome of other Brachionus rotifers have been published from Brachionus plicatilis, Brachionus koreanus, Brachionus rotundiformis, Brachionus calyciflorus, Brachionus paranguensis, and Brachionus rubens (Suga et al. Citation2008; Hwang et al. Citation2013, Citation2014; Kim et al. Citation2017; Choi et al. Citation2019; Choi, Kim, et al. Citation2020; Choi, Lee, et al. Citation2020). Thus, the revealing of complete mitochondrial genome of B. angularis would be helpful to better understand the phylogenetic relationship of B. angularis species complex clade. Also, B. angularis is considered as a model for aquaculture (Ogata et al. Citation2011; Ogello and Hagiwara Citation2015; Ogello et al. Citation2016), environmental biology (Ferrão-Filho et al. Citation2002; Wang et al. Citation2016), and ecology (Yang et al. Citation2009; Zhang et al. Citation2010; Yin et al. Citation2017) in response to environmental factors. In this study, we identified two complete mitochondrial genomes of the monogonont rotifer B. angularis.
The resting eggs of B. angularis were collected by Dr. E.O. Ogello (Kenya Marine and Fisheries Research Institute in Kenya) by netting from sediments of freshwater ponds (0°42′35.7′′S and 34°49′11.3′′E) in August 2014, then transported to the Laboratory of Aquaculture Biology, Nagasaki University, Japan for further study (Ogello et al. Citation2016). To identify complete mitochondrial DNA of B. angularis, the live samples were sent to South Korea. The type of B. angularis (85.6 μm in length and 75.4 μm in width) was deposited at the Ichthyological collection of the Faculty of Fisheries, Nagasaki University (FFNU) under the accession no. FFNU-Rot-0006.
We sequenced B. angularis from whole body genomic DNA using the nanopore platform (Oxford Nanopore Technologies, Oxford, United Kingdom). De novo assembly was conducted by smartdenovo (https://github.com/ruanjue/smartdenovo). For the assembled B. angularis 106 contigs (63,578,663 bp), Pilon version 1.23 (https://github.com/broadinstitute/ pilon/releases) and the 300 bp HiSeq2500 (Illumina, San Diego, CA) data were employed for polishing processes, and obtained one complete mitochondrial DNA sequence (mitochondrial DNA I) through manual editing process. In addition, to identify the second complete mitochondrial genome (mitochondrial DNA II), we revisited and examined the initially assembled 134,733 contigs (80,281,057 bp), generating from de novo assembly of 300 bp HiSeq2500 (Illumina) data with Newbler version 2.9 (http://www.454.com), based on the complete mitogenome of the freshwater rotifer B. rubens.
The complete mitochondrial genomes of B. angularis were 10,764 bp (mitochondrial DNA I; GenBank no. MT875425) and 12,238 bp (mitochondrial DNA II; GenBank no. MT875426) in size. The gene structure and its orientation of 12 PGCs of complete mitochondrial genomes of B. angularis were identical to those shown in other marine rotifers and the freshwater rotifer B. rubens, but was different from the freshwater rotifer B. calyciflorus that had a different combination of 12 PGCs with additional cytochrome b gene in the mitochondrial DNA I. Of 12 protein-coding genes (PCGs), one gene (ND5) had incomplete stop codon. Furthermore, GTG was identified as the start codon for CO1, ND4L, ND5, and CO2 while ATA was the start codon for ND3, whereas the start codon for other PCGs was ATG. The base composition of 12 PCGs in B. angularis mitogenome showed 20.4% for A, 47.3% for T, 17.5% for C, and 14.8% for G, respectively. The mitochondrial genome A + T base composition (67.7%) of 12 PCGs was higher than G + C (32.3%), whereas the complete mitochondrial genome A + T base composition (66.3%) was higher than G + C (33.7%).
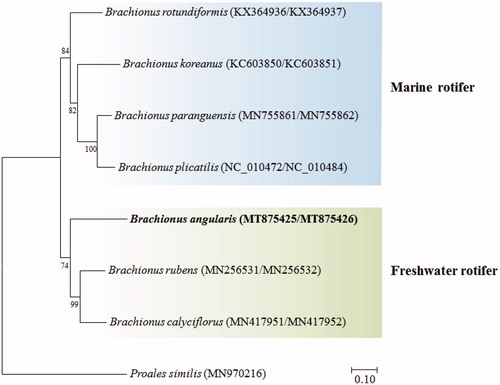
The placement of B. angularis in the genus Brachionus with 12 PGCs was shown in . B. angularis was clustered with B. rubens and B. calyciflorus which are freshwater species, but was clearly separated from the marine species such as B. rotundiformis, B. koreanus, and B. paranguensis, possibly suggesting relationship between the differences in their natural habitat and mitogenome.
Figure 1. Phylogenetic analyses based on mitochondrial DNA of Brachionus angularis with seven congeners. The amino acid sequences of 12 mitochondrial DNA genes were aligned by ClustalW. Maximum likelihood analysis was performed by Mega software (version 10.0.1) with Gamma + LG + I model. The rapid bootstrap analysis was conducted with 1000 replications with 48 threads running in parallel. The rotifer Proales similis (class Monogononta) served as outgroup. −Ln = 28545.407996.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference numbers MT875425 and MT875426.
Additional information
Funding
References
- Choi BS, Kim DH, Lee JS, Kim HJ, Hagiwara A, Lee JS. 2020. Complete mitochondrial genome of the euryhaline monogonont rotifer Brachionus paranguensis (Rotifera, Brachionidae). Mitochondrial DNA Part B. 5(1):502–503.
- Choi BS, Lee YH, Hagiwara A, Lee JS. 2019. Complete mitochondrial genome of the freshwater monogonont rotifer Brachionus calyciflorus (Rotifera, Brachionidae). Mitochondrial DNA Part B. 4(2):3593–3595.
- Choi BS, Lee YH, Lee JS, Ogello EO, Kim HJ, Hagiwara A, Lee JS. 2020. Complete mitochondrial genome of the freshwater monogonont rotifer Brachionus rubens (Rotifera, Brachionidae). Mitochondrial DNA Part B. 5(1):5–6.
- Ferrão-Filho AS, Kozlowsky-Suzuki B, Azevedo SMFO. 2002. Accumulation of microcystins by a tropical zooplankton community. Aquat Toxicol. 59(3–4):201–208.
- Hu H, Xi Y. 2008. Demographic parameters and mixis of three Brachionus angularis Gosse (Rotatoria) strains fed on different algae. Limnologica. 38(1):56–62.
- Hu HY, Xi YL, Geng H. 2003. Comparative studies on individual growth and development of three Brachionus angularis strains. Chinese J Appl Ecol. 14:565–568.
- Hwang DS, Dahms HU, Park HG, Lee JS. 2013. A new intertidal Brachionus and intrageneric phylogenetic relationship among Brachionus as revealed by allometry and CO1-ITS1 gene analysis. Zool Stud. 52(1):13.
- Hwang DS, Suga K, Sakakura Y, Hagiwara A, Park HG, Rhee JS, Lee JS. 2014. Complete mitochondrial genome of the monogonont rotifer, Brachionus koreanus (Rotifera, Brachionidae)). Mitochondrial DNA. 25(1):29–30.
- Kim HS, Hwang DS, Kim HJ, Sakakura Y, Hagiwara A, Lee JS. 2017. Complete mitochondrial genome of the monogonont rotifer Brachionus rotundiformis (Rotifera, Brachionidae). Mitochondrial DNA Part B. 2(1):39–40.
- Ogata Y, Tokue Y, Yoshikawa T, Hagiwara A, Kurokura H. 2011. A Laotian strain of the rotifer Brachionus angularis holds promise as a food source for small-mouthed larvae of freshwater fish in aquaculture. Aquaculture. 312(1–4):72–76.
- Ogello EO, Hagiwara A. 2015. Effects of chicken manure extract on the population growth, mixis induction and body size of the freshwater rotifer Brachionus angularis Gosse 1851. Asian Fish Sci. 28:174–185.
- Ogello EO, Kim HJ, Suga K, Hagiwara A. 2016. Life table demography and population growth of the rotifer Brachionus angularis in Kenya: influence of temperature and food density. African J Aquat Sci. 41(3):329–336.
- Segers H. 2007. Annotated checklist of the rotifers (Phylum Rotifera), with notes on nomenclature, taxonomy and distribution. Zootaxa. 1564(1):1–104.
- Suga K, Mark Welch DB, Tanaka Y, Sakakura Y, Hagiwara A. 2008. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol Biol Evol. 25(6):1129–1137.
- Wang C, Wang L, Deng D, Zhou Z. 2016. Temporal and spatial variations in rotifer correlations with environmental factors in Shengjin Lake, China. Environ Sci Pollut Res. 23(8):8076–8084.
- Yang G, Zhong C, Pan H. 2009. Comparative studies on seasonal variations of metazooplankton in waters with different eutrophic states in Lake Taihu. Environ Monit Assess. 150(1–4):445–453.
- Yin X, Jin W, Zhou Y, Wang P, Zhao W. 2017. Hidden defensive morphology in rotifers: benefits, costs, and fitness consequences. Sci Rep. 7(1):4488.
- Zhang S, Zhou Q, Xu D, Lin J, Cheng S, Wu Z. 2010. Effects of sediment dredging on water quality and zooplankton community structure in a shallow of eutrophic lake. J Environ Sci. 22(2):218–224.
