Abstract
Vatica guangxiensis S.L. Mo is an evergreen large tree of Dipterocarpaceae. Herein, we assembled the complete chloroplast genome of Vatica guangxiensis by next-generation sequencing technologies. The complete chloroplast genome sequence of Vatica guangxiensis is 151,010 base pairs (bp) in length, including a pair of inverted repeat regions (IRs, 23,827 bp), one large single-copy region (LSC, 83,353 bp), one small single-copy region (SSC, 20,003 bp). Besides, the complete chloroplast genome contains 123 genes in total, including 83 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that Vatica guangxiensis has the closest relationship with Vatica mangachapoi. Our study lay a foundation for further research of Vatica mangachapoi.
Vatica guangxiensis is a characteristic tree species of tropical rain forest in Southern Yunnan of China and it is also an important timber tree species (Pachynocarpus and Retinodendron Citation2007). This species has limited number of individuals, only with three natural populations distributed in Southern Yunnan and Liushaoshan in Napo County of Guangxi, China. It was listed as an endangered plant species in China (Li et al. Citation2002). Mass extinction that is primarily due to large-scale habitat destruction caused by human activities. The best remedy to prevent extinction is habitat preservation. With the over-excavation of Vatica guangxiensis, their habitats have been broken up and the wild resources have decreased significantly over the last decades. Therefore, the Vatica guangxiensis is listed as an endangered species in the China Species Red List (Wang and Xie Citation2004). Therefore, we report the complete chloroplast genome of Vatica guangxiensis, in order to better understand the relationship between Vatica guangxiensis and related genera, and contribute to the effective conservation strategy of Vatica guangxiensis.
The sample of Vatica guangxiensis. was collected from Yachang Orchidaceae National Nature Reserve, Guangxi, China (24°44′N, 106°15′E) and the voucher specimen deposited at Herbarium of Guangxi Institute of Botany, Guangxi Zhuang Autonomous Region and Chinese Academy of Sciences (specimen code Guangxiensis _ GX).
High-quality genomic DNA of Vatica guangxiensis was extracted from leaves by TIANGEN plant genomic DNA kit, and sequenced by the BGISEQ-500 platform. With the chloroplast genome of Vatica mangachapoi (GenBank accession number NC_041485) as the reference sequences, we assembled the complete chloroplast genome from the clean reads by the GetOrganelle pipe-line (Jin et al. Citation2018), and then annotated the new sequences using the Geneious R11.15 (Kearse et al. Citation2012). Finally, a complete chloroplast genome of Vatica guangxiensis was obtained and submitted to Genbank (accession number MT934442).
The complete chloroplast genome sequence of Vatica guangxiensis is 151,010 base pairs (bp) in length, including a pair of inverted repeat regions (IRs, 23,827 bp), one large single-copy region (LSC, 83,353 bp), one small single-copy region (SSC, 20,003 bp). Besides, the complete chloroplast genome contains 123 genes in total, including 83 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. In addition, the overall GC content of the genome was 37.2%.
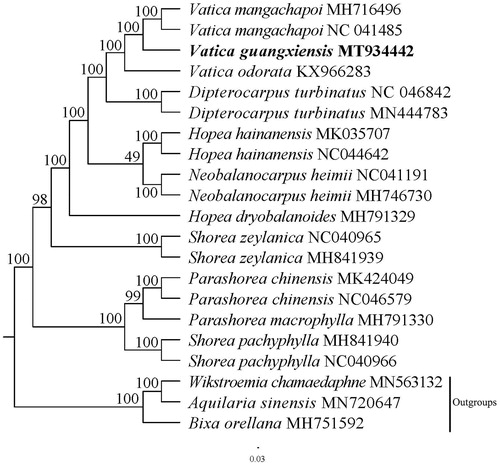
In order to confirm the phylogenetic position of Vatica guangxiensis, a maximum likelihood analysis was performed by MEGA 6.0 (Tamura et al. Citation2013) with 1000 bootstrap replicates (Minh et al. Citation2013; Chernomor et al. Citation2016) based on 21 complete chloroplast genomes. All sequences were aligned with the HomBlock pipeline (Bi et al. Citation2018) and subsequently checked manually in Bioedit v5.0.9 (Hall Citation1999). The results showed that Vatica guangxiensis was sister to Vatica mangachapoi with 100% bootstrap support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data openly available in a public repository that does not issue DOIs. The data that support the findings of this study are openly available in [National Center for Biotechnology Information] at [https://www.ncbi.nlm.nih.gov/], reference number [MT934442].
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Sym-Posium Series. 41:95–98.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv, 256479
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li QM, Xu ZF, He TH. 2002. A preliminary study on conservation genetics of endangered Vatica guangxiensis (Dipterocarpaceae). Acta Botanica Sinica. (02):246–249.
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 30(5):1188–1195.
- Pachynocarpus JDH, Retinodendron K. 2007. Flora of China. Vol. 13. Dipterocarpaceae. Beijing: Science Press, p. 52–54.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30(12):2725–2729.
- Wang S, Xie Y. 2004. China species red list. Vol. 1: RedList. Beijing: Higher Education Press, p. 367.